Ocean robots uncover microbial secrets
Issue: Oceans
05 February 2019 article

Life on Earth began in the sea, and the oceans continue to support life on our planet. Of particular importance is the ability of marine microbes to exist in a complex web of relationships where substances are continually transformed and exchanged.
For instance, some microbes take energy from the sun and convert carbon dioxide into biomass, while other microbes consume this biomass, releasing carbon dioxide into the atmosphere. This is an example of a biogeochemical ‘cycle’ that is central to all life on the planet, yet understanding how these microbes perform this cycling requires observation capabilities that are not yet fully developed. This is because understanding biogeochemical cycling in the ocean requires samples, taken at times and locations more frequently than the microbial signature is changing. This is a difficult proposition for typical ship-based sampling schemes.
Traditional oceanic sampling techniques involve deploying collection bottles from scientific research vessels. Once these bottles are onboard, the seawater is filtered and these filters are either processed aboard a ship or preserved for laboratory processing on shore. Sampling difficulties related to ship operations, ship cost, time-on-station and weather, can all impact how many samples humans aboard a ship can collect and process. The use of autonomous samplers overcomes many of these difficulties, by allowing a more persistent presence in the environment of interest, unaffected by weather or ship schedules. This persistent presence allows sampling at higher spatiotemporal scales (e.g. hourly and over several depths/days), which may provide insights into biogeochemical cycles that are hidden in the low-frequency sampling of ship expeditions. Many autonomous samplers can also be integrated into gliders or autonomous underwater vehicles (AUVs), allowing instruments to drift with ocean currents or sample in remote regions. Here we provide an overview of three novel autonomous sampling devices that can be useful in molecular ecological studies, including the newer ‘omics’ investigations (metatranscriptomics, proteomics: or holistic RNA and protein studies).
MAPS and N2 fixers
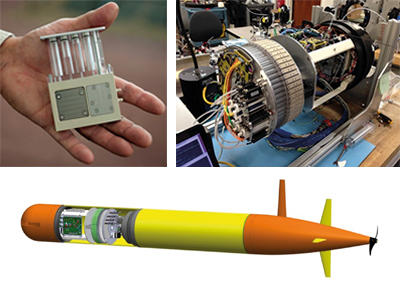
The Marine Autonomous Plankton Sampler (MAPS) is an autosampler developed by the National Oceanography Centre, Southampton. MAPS was designed to acquire planktonic biomass by filtering large volumes of seawater through commercially available cartridge filters, allowing cells > 0.2 µm in size to be collected and preserved. MAPS can filter and archive a two-litre open ocean sample every 30 minutes (Fig. 1).
As a major limiting nutrient in the ocean, nitrogen controls the magnitude of atmospheric CO2 uptake and fuels the base of the food web in many marine environments. In addition to anthropogenic sources, fixation of N2 gas by microbes can contribute to bioavailable nitrogen and primary production. As N2-fixing microbes are typically patchy in distribution, the use of autonomous samplers is important in advancing marine N2 fixation research.
During a cruise in 2017, MAPS was used to characterise and quantify the activities of marine N2-fixing microbes. Hourly samples were collected over the 10 days across a range of high- and low-biomass regimes for metatranscriptomics and DNA analysis. Quantification of genes by quantitative PCR reported similar abundances relative to flash freezing, supplying datasets on micro-organisms in the North Atlantic. Now that MAPS has been proven in the field, it will be validated across a range of platforms in order to address needs in the fields of conservation (environmental DNA), biogeochemistry and aquaculture.
The ESP and ocean eddies
The Environmental Sample Processor (ESP) is an autonomous in situ water sample collection and processing device developed by Monterey Bay Aquarium Research Institute (MBARI). Using an electromechanical fluidic system, water samples are autonomously collected and filtered, and filters are then either preserved and stored, or processed on board. In situ analytics include array hybridisation with nucleic acid probes or quantitative PCR. A new generation of ESP was recently developed to be much smaller and fit within an MBARI-developed vehicle, the Long-Range AUV (LRAUV; Fig. 2), in order to perform high-frequency sampling within large, open-ocean oceanographic eddies.
These rotating water masses can be hundreds of km in diameter and are hotspots of microbial life due to the fact that the slow rotation of an eddy can bring cold, nutrient-rich water into surface layers from deeper depths. In these eddies, phytoplankton typically concentrate around 100–120 metres deep, in a layer known as the deep chlorophyll maximum. Understanding how eddies affect the deep chlorophyll maximum and influence ocean productivity is currently not well understood because of difficulties sampling these mobile features.
The ESP-LRAUV system was recently deployed north of Maui, Hawaii, in collaboration with the University of Hawaii’s Simons Collaboration on Ocean Processes and Ecology (SCOPE) programme to study the microbial community directly associated with eddies. In particular, researchers hope to understand how gene activation in marine microbes changes through a full day/night cycle, and how these changes ultimately affect the productivity of these ubiquitous eddy communities. The ESP-LRAUV found the deep chlorophyll maximum at the centre of an eddy, and began sampling every three hours for three full days, all while maintaining position at 100 metres depth, and within the eddy centre as the eddy slowly drifted west. Samples collected every three hours were preserved and returned to the laboratory for processing once the vehicle was recovered. Deploying this novel technology allows microbial samples to be collected within important features of the Earth’s ocean circulation that have been difficult to sample repeatedly over a day/night cycle.
Clio and marine microbial mapping
Clio is an AUV developed by Woods Hole Oceanographic Institute (WHOI) and the University of Texas Rio Grande Valley (UTRGV). It was explicitly designed for ocean mapping of marine microbial communities and metabolic biomarkers (Fig. 3), and to reduce sampling time during vertical surveys from the surface to seafloor. Clio is a filtering and sampling machine the size of a large refrigerator. When deployed from a vessel, Clio can dive independently to a maximum depth of 6,000 metres and return within hours, collecting and preserving samples along the way. Hundreds of litres of seawater are filtered for each sample, allowing a wide range of biochemical measurements to be made from the particulate samples collected, even from depths where particulate matter is scarce. Clio significantly reduces the time required to collect these water column profiles, and the goal is that this increased sampling efficiency will facilitate a more thorough and rapid mapping of ocean biochemical processes. Clio also collects biomass samples suitable for omics investigations, allowing researchers to answer questions about the response of marine microbes to environmental stressors.
Previous global studies have mapped ocean structure and the distribution of major marine nutrients. However, a co-ordinated mapping of marine microbial community structure and markers of biochemical function has yet to be undertaken; though there is considerable potential and excitement about this possibility. A global dataset of marine microbial community composition and relevant biochemistry would be of tremendous value for improving and validating marine ecosystem and biogeochemical models.

Clio has been operational for a little over a year, and is being used in a study analysing the nutrient stress response in cyanobacteria communities in the Atlantic Ocean. As part of this study, it is being used for repeat profiles at the Bermuda Atlantic Time-series Study station until next summer when it will be used to create a sectional map of nutrient stress biomarkers from Bermuda to the North American continental shelf.
Future prospects
High-resolution autonomous sampling has already resulted in the discovery of ‘pulses’ of microbial activities that weren’t previously predicted based on direct assessments of microbial physiology or in cultured representatives, as well as the validation of predictions resulting from nutrient controls and thermodynamics-based modelling in the deep sea. Coming years will yield more discoveries using ocean robots, giving microbial oceanographers new eyes to observe change in this complex environment. We envision that broader global omics observations will deliver a new understanding, leading to prediction of marine microbial dynamics in the coming decades.
Further reading
Anantharaman K, Breier JA, Dick GJ. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J 2016;10:225.
Birch J, Barone B, DeLong E, Foreman G, Gomes K. et al. Autonomous targeted sampling of the deep chlorophyll maximum layer in a subtropical North Pacific eddy. OCEANS 2018 MTS/IEEE Charleston . Charleston, SC, USA:IEEE;2018, pp. 1–5.
Breier JA, Sheik CS, Gomez-Ibanez D, Sayre-McCord RT, Sanger R, et al. A large volume particulate and water multi-sampler with in situ preservation for microbial and biogeochemical studies. Deep Sea Res Part 1 Oceanogr Res Pap 2014; 94:195–206.
Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R 3rd. Ocean microbes. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science 2014;345:207–212.
Robidart JC, Magasin JD, Shilova IN, Turk-Kubo KA, Wilson ST et al. Effects of nutrient enrichment on surface microbial community gene expression in the oligotrophic North Pacific Subtropical Gyre. ISME J DOI:10.1038/s41396-018-0280-0 [Epub ahead of print]
Robidart J, Wyatt J, Brown R, Walk J, Cerdán E et al. Design, development and validation of the Marine Autonomous Plankton Sampler for high-resolution collection and preservation of microbes and eDNA. Environ Sci Technol, unpublished.

Susan Evans
Ocean Technology and Engineering, The National Oceanography Centre (NOC), Southampton SO14 3ZH, UK
[email protected]
Twitter: @SusanKEvans
Susan Evans is a microbial biogeochemist. Her research currently focuses on developing a DNA/RNA extraction chip and the early detection of harmful algae in shellfish farms using miniaturised impedance cytometry.

Jim Birch
Monterey Bay Aquarium Research Institute, CA, USA
Jim Birch is the director of the SURF Center (Sensors: Underwater Research of the Future) where he oversees all aspects of the Environmental Sample Processor (ESP) project.

John 'Chip' Breier
University of Texas Rio Grande Valley, TX, USA
John 'Chip' Breier is a marine geochemist and ocean engineer who studies the coupling between biotic and abiotic processes, from the seafloor to the surface ocean.

Michael Jakuba
Woods Hole Oceanographic Institution (WHOI), MA, USA
Michael Jakuba is a Senior Engineer in the Applied Physics and Ocean Engineering Department at WHOI, who develops underwater robotic systems in support of oceanographic science.

Mak Saito
Woods Hole Oceanographic Institution (WHOI), MA, USA
Mak Saito is a Senior Scientist in the Marine Chemistry and Geochemistry Department at WHOI, who studies micronutrient use in marine microbes and their influence on biogeochemical cycles, using proteomics and metal analyses.

Julie Robidart
Ocean Technology and Engineering, The National Oceanography Centre (NOC), Southampton SO14 3ZH, UK
Julie Robidart is a molecular ecologist in Ocean Technology and Engineering, leading a biosensor programme focused on the development and application of novel technologies for analysing marine microbes. Her research involves analysis of environmental metagenomic and metatranscriptomic datasets to inform probe design and development and optimisation of molecular assays for autonomous instrumentation.
Why does microbiology matter?
Susan: Microbes play a key role in maintaining life on Earth. Roughly half the oxygen produced on the planet every day is made by micro-organisms that live in the ocean!
Julie: We wouldn't be here if it weren't for microbes making our planet hospitable!
What is your greatest achievement to date?
Susan: Submitting my PhD thesis on dimethylsulphoxide (DMSO) utilising bacteria in the ocean. I really enjoyed my project but there were many challenges along the way.
Julie: Learning to navigate academia and what to prioritise, in the old days before early career training was ubiquitous.
Fig. 1. (thumbnail): MAPS Marine Autonomous Plankton Sampler on the long-term buoy L4 of the Western Channel Observatory, English Channel. James Fishwick, Plymouth Marine Laboratory .
Fig. 2. The latest ESP design is based on cartridges (upper left), 60 of which are loaded around a central ring
(upper right). The lower panel shows how the ESP fits within the LRAUV payload. Jim Birch, MBARI.
Fig. 3. Clio, a biochemical mapping autonomous vehicle, preparing for launch from the R/V Atlantic
Explorer at the Bermuda Atlantic Time-series Study station. UTRGV.


