Genome segregation in heat-loving archaea
Issue: Archaea
08 August 2017 article
Archaea are remarkable objects of investigation due to their exquisitely distinctive biological properties and macromolecules. Since their discovery four decades ago, there has been an escalation in knowledge, genome sequences and publications on this domain of life. However, the fundamental process of genome segregation remains a terra still vastly incognita in this branch on the sprawling tree of life.
Archaea are single-cell organisms that populate planet Earth and which together with bacteria and eukaryotes form the three domains of life. Archaea appear similar to bacteria, but belong to a different branch of the tree of life. Both bacteria and archaea are prokaryotes, i.e. their genetic material is not wrapped by a membrane into a separate compartment, called a nucleus, which instead is a hallmark of eukaryotes such as baker’s yeast, fungi, plants and animals, including humans to mention some.
Archaea are ubiquitous and constitute a considerable fraction of the global biosphere. Some live ordinary lives in mundane environments, such as lakes, seas and insect intestinal tracts, whereas others live extraordinary lives pushed to extremes in incredibly harsh habitats, such as hot springs, deep-sea hydrothermal vents, volcanic mud and salt lakes. Archaea have been instrumental in evolutionary studies on the origin of life and have revealed to us that the boundaries of life as we know it can be pushed much further than previously anticipated.
Thermophilic or heat-loving archaea
Thermophilic archaea are super microbes that thrive at 80 ºC and higher temperatures in hot springs, volcanoes and deep-sea vents. These archaea exhibit fascinating properties, which make them valuable for the development of novel biotechnological applications, but also extremely interesting for basic studies on life pushed to extremes. Thermophilic archaea contain heat- stable cellular building blocks such as proteins and lipid chains in the cell membrane that represent adaptations to the habitats in which these superbugs thrive. The ability of these archaea to grow in extreme environments where no other terrestrial organism can survive has also rekindled hopes of discovering extraterrestrial life on similarly inhospitable planets of our solar system as well as in more far-flung reaches of the universe.
Genome segregation
Genome segregation is a crucial stage of the life cycle of every cell: the DNA is first duplicated, then separated and equally distributed into the two daughter cells. Despite the significant progress made in decoding molecular mechanisms in archaea in the last four decades, a paucity of information is available to date on the fundamental process of DNA segregation in these micro-organisms. The subject remains a black box awaiting investigation. Nevertheless, recent insights into this process have emerged from investigations on species of the heat-loving archaeon genus Sulfolobus that have been isolated from acidic hot springs and solfatara fields across the planet.
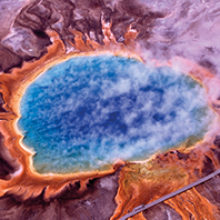
Microscopy images of Sulfolobus solfataricus cells, bright field (left) and overlay with DAPI-stain (blue chromosome) (right). Bar, 4.5 μm.
Sulfolobus solfataricus and the SegAB toolkit for chromosome segregation
The late German scientist Wolfram Zillig was an archaea pioneer. Zillig travelled from Germany to Naples in 1978 to accomplish a mission: re-isolating a thermophilic archaeon from the Earth’s viscera in an area west of Naples known as Campi Flegrei or ‘Burning Fields’. Campi Flegrei is a large volcanic region harbouring a caldera that is basically at ground level. It is possible to walk on the caldera terrain, which is dotted with fumaroles that emit sulfurous steam and with many pools of boiling mud. It is a truly infernal landscape. From this site, Zillig isolated an archaeal species that he called Sulfolobus solfataricus. This archaeon is a strict aerobe that grows at 80 ºC and a highly acidic pH.
The genome sequence of S. solfataricus was determined in 2001. The sequence revealed two genes potentially involved in chromosome segregation, i.e. the separation and delivery of the newly replicated sister chromosomes to daughter cells at the stage of division. The genes were later designated as segA and segB for chromosome segregation. The SegA protein is a cousin of bacterial factors called ParA, which are adenosine triphosphate (ATP)-binding and -hydrolysing enzymes encoded by low-copy-number plasmids and chromosomes, and whose function is crucial for accurate genome segregation. The SegB protein is an archaea-specific factor that does not show sequence similarity to any eukaryotic or bacterial protein.
Recent investigations have shown that SegA is indeed an ATP-hydrolysing protein that assembles into higher-order structures in vitro upon ATP binding, whereas SegB is a site-specific DNA-binding protein that recognises DNA sequences located upstream of the segAB genes. The two proteins assemble into a complex. Furthermore, SegB plays a role in dictating the accretion of SegA multi-subunit structures. The fact that SegB is attracted to specific DNA sequences and becomes glued to them is important, as the entire complex is recruited to those chromosomal locations. These sites are believed to be key to the separation of the chromosomes. When the number of subunits of SegA and SegB proteins in S. solfataricus cells is increased by using some genetic trickery, chromosome segregation is disrupted as shown by microscopy investigations, which reveal cells without chromosomes, others with the chromosomal DNA squeezed into one-half of the cell volume and some others with guillotined chromosomes. These defects indicate that the SegA-SegB complex plays a vital role in chromosome segregation. The SegA-SegB proteins are believed to separate and drive apart sister chromosomes, although the mechanism underpinning this process remains to be elucidated. The way one can picture this system is as a highly precise DNA molecule sorting machine.
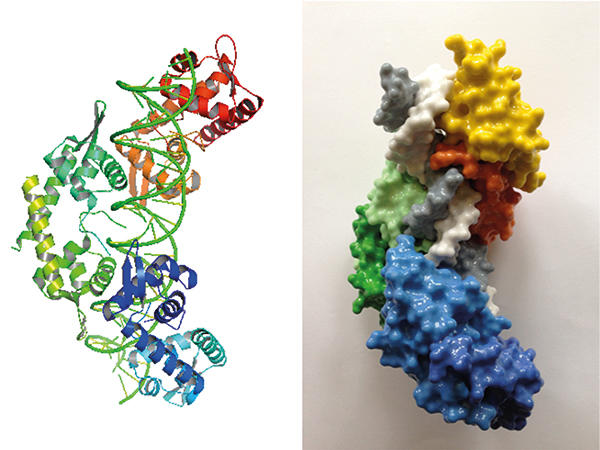
Structure (left) and 3-D printed model (right) of pN0B8 AspA-DNA complex showing three dimers (orange, green and blue) associated with DNA (white and grey).
Borrowing building blocks from Bacteria and Eukarya: a three-component plasmid segregation machine in Archaea
Another story dates back to the 1990s and begins with a further mission of Wolfram Zillig and coworkers, who travelled to the island of Hokkaido, Japan. From acidic hot springs at Noboribetsu in Hokkaido, Zillig isolated a Sulfolobus strain containing a plasmid, pNOB8, that harbours three genes encoding an atypical DNA segregation system.
One of the segregation proteins encoded by pNOB8, AspA, is a site-specific DNA-binding protein. AspA recognises an inverted repeat sequence on pNOB8 and then associates to adjacent regions, thereby spreading on the DNA on which the protein forms a left-handed helix. ParA and ParB are the other two proteins of this plasmid-sorting machine. ParB is an adaptor that sits between AspA and ParA within the complex. Adaptors need to be pliable, and ParB is indeed flexible: the protein consists of two domains connected by an extended flexible linker. One of the ParB domains has a bacterial flavour; whereas the other resembles CenpA, which is a eukaryotic histone that replaces the canonical histone H3 at centromeres and is crucial for assembly of the kinetochore complex that drives chromosome pairs apart in eukaryotic cells. Structural investigations have shown that the AspA helix functions as a docking platform onto which ParB molecules assemble into a second superhelix that fits into the AspA-DNA structure in a lock-and-key fashion. In a nutshell, ParB subunits form a ‘roller coaster’ that is anchored on the AspA spiral, a truly remarkable assembly. ParA is then recruited into the complex via interaction with ParB and potentially mediates plasmid anchoring to the chromosome through its nonspecific DNA-binding activity. The mechanism that underpins pNOB8 partition remains under investigation.
The AspA-ParB-ParA multi-protein complex in archaea merges bacterial and eukaryotic features suggesting the possible conservation of DNA segregation principles across the three domains of life. Therefore, studies on archaea are providing new and intriguing perspectives on genome segregation that are pertinent to all life on planet Earth.
Acknowledgements
The work on genome segregation in DB laboratory is funded by the BBSRC and Leverhulme Trust.
Daniela Barillà
Department of Biology, University of York, York YO10 5DD
[email protected]
@daniela_barilla
Further reading
Barillà, D. (2016). Driving apart and segregating genomes in Archaea. Trends Microbiol 24, 957–967.
Kalliomaa-Sanford, A. K. & others (2012). Chromosome segregation in Archaea mediated by a hybrid DNA partition machine. Proc Natl Acad Sci U S A 109, 3754–3759.
Schumacher, M. A. & others (2015). Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages. Science 349, 1120–1124.
Images: Aerial view of the Grand Prismatic Spring, Midway Geyser Basin, Yellowstone National Park. Sulfolobus species have been isolated from acidic hot springs in Yellowstone. NPS photo by Jim Paeco. Microscopy images of Sulfolobus solfataricus cells. Brett McLeod. Structure and 3-D printed model of pN0B8 AspA-DNA complex. D. Barillà.


