Editorial
Issue: SARS-CoV-2 and COVID-19
12 October 2021 article
Welcome to the October 2021 issue of Microbiology Today! As I write this Editorial in late August, many of us are preparing ourselves for working through a period of yet more uncertainty and volatility as we continue to learn how to deal with the ongoing presence (and consequences) of SARS-CoV-2 in our day-to-day lives. Indeed, as we approach the end of our second year of the COVID-19 pandemic, we have decided to make SARS-CoV-2 and COVID-19 the sole focus of this issue.
I feel that I am understating the matter when I say that SARS-CoV-2 and COVID-19 have had a profound impact on our professional and personal lives. Yet despite this, we have all made incredible efforts to adapt to pandemic life and as professional microbiologists we have shown a captive audience once again why microbiology matters. Many of us have contributed to understanding the virus at a molecular and clinical level, developed diagnostics, treatments and vaccines, and communicated all these aspects and many more to the general populace so they can understand what is happening in the world around them. It is this work that forms the focus of our October issue.
Keith Grehan sets the scene for our issue by providing an overview of the molecular virology of SARS-CoV-2. Keith distils a wealth of information to describe the key features of the SARS-CoV-2 genome. An ongoing concern is the emergence of SARS-CoV-2 variants with altered characteristics. Keith concludes by discussing the consequences of key amino acid substitutions found in several variants of concern.
In the present day we seem to be inundated with requests to have a ‘COVID test’, and in the UK any suspicion of COVID-19 results in the rapid deployment and turnaround of PCR-based testing. This infrastructure was almost non-existent in early 2020, and in our next article Alan McNally provides us with a fascinating personal insight into these early days of setting up large-scale SARS-CoV-2 testing in the UK. As Alan notes in his article, the advantage of incorporating genomics into infectious disease diagnostics has been highlighted in this epidemic, particularly in using the data generated to monitor the emergence and spread of new variants. Colman O’Cathail expands on this area in his own perspective, by describing the ongoing work of bioinformaticians to rapidly archive and make publicly available the vast quantities of SARS-CoV-2 sequencing data generated around the world.
As an increasing number of COVID-19 cases appeared around the world, we heard of infected individuals suffering from a loss of taste. As these anecdotes gathered an evidence base and became incorporated into criteria for a presumptive diagnosis of COVID-19, work began to understand the mechanistic basis behind the emergence of these symptoms. David Edwards and Alexander Gardner take us through this body of work in our next article, discussing amongst other things the potential for SARS-CoV-2 to replicate within cells of the oral cavity.
Whilst efforts were underway early in the pandemic to develop vaccines effective in preventing infection, there was a parallel need to find therapeutics capable of treating infection. In our fifth article, Jane Hilton and Catherine Adamson review the challenges faced both in identifying existing agents for repurposing into COVID-19 treatments and in identifying new targets to exploit in the development of new agents.
As we approach the second anniversary of the emergence of COVID-19, our final two articles look at what the future may hold for SARS-CoV-2, and the lessons we can apply to future pandemics. Grace Roberts looks at the challenges of communicating SARS-CoV-2 research to the public, with a focus on perceptions surrounding the introduction of face coverings and the rollout of vaccines. Tim Inglis, who has contributed coronavirus blogs to the Society for most of the pandemic, draws the October issue to a close with his commentary on the future of COVID-19 and the impact of policy and health infrastructure in individual countries on the persistence of this pandemic.

Chris Randall
Editor
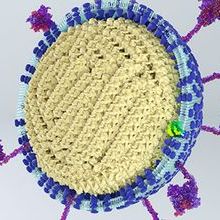
Cover image: This image is from a collaboration between Annabel Slater, who specialises in science communication and visualisation, and Ed Hutchinson, a virologist at the MRC-University of Glasgow Centre for Virus Research. SARS-CoV-2 virions are irregular structures that cannot be readily resolved by any one method, and this model brings together multiple lines of information about SARS-CoV-2 and other related coronaviruses to provide a clear visualisation of their complex structure.
Image: Annabel Slater/MRC-University of Glasgow Centre for Virus Research.


