How can you increase the rigour and transparency in your research? Submit to our open research platform
Issue: SARS-CoV-2 and COVID-19
12 October 2021 article
Over the past year during the pandemic, there has been a complete overhaul of the way research is undertaken and shared. Researchers need to rapidly share their work, often before publication, and are increasingly being required to share the underlying results of their research too.
This has put the open science agenda at the forefront for the Microbiology Society, and we recognised the need for a trusted place for researchers to disseminate their work rapidly, rigorously and transparently. Using a grant awarded by the Wellcome Trust and the Howard Hughes Medical Institute (HHMI), the Microbiology Society is converting our sound science, Open Access journal, Access Microbiology, into an open research platform. The platform will launch and be open for submissions at the beginning of 2022.
How will the platform work?
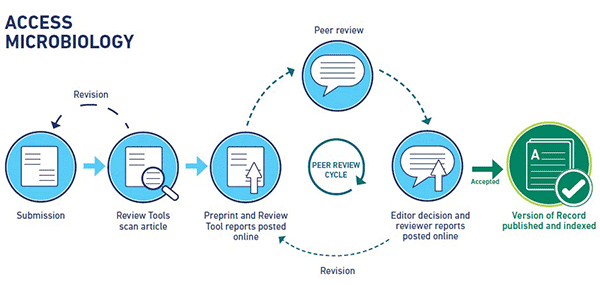
The Access Microbiology open research platform will become a home for the entire life cycle of an article, from the author’s very first preprint all the way to the published Version of Record. Authors will submit to our peer review system as usual and will be given the opportunity to improve their article using feedback from three manuscript review tools, Penelope.ai, SciScore and iThenticate. Once the authors are happy with their article and it complies with the platform’s ethical and editorial policies, the article is posted online as a preprint with a digital object identifier (DOI), giving authors the opportunity to start to receive community feedback on their work. The reports from the manuscript review tools will also be posted alongside the preprint, allowing readers to perform their own ‘health check’ on the preprint prior to completing peer review. In an effort to increase the rigour and reproducibility of the research, the platform will adopt an open data policy, so all articles will include links to open data, code and methods, allowing readers to access the underlying data of the work.
Meanwhile, the preprint is assigned to an Editorial Board Member (EBM) and will undergo full, transparent peer review. We recognise that reviewers might not always want to put their name to their review, so whilst the content of all reviews will be posted alongside the article, reviewers can choose to remain anonymous if they wish. Once the Editor makes their recommendation, the reviewer reports and Editor’s decision and comments will be posted online. Reviewers can therefore receive public recognition of their work, if they wish, and readers can assess these reviews themselves, promoting trust in the publishing process. Articles that are considered acceptable for publication will then be fully typeset in our platform style and branding, published on the platform in full-text HTML and PDF, and deposited in PubMed, the open access repository PubMed Central, and other indexing services.
What are the benefits over traditional journal publishing?
There are various unique and innovative aspects to the platform that mean authors will benefit more than if they were to submit to a normal preprint server and journal. Firstly, during submission authors can choose to receive immediate feedback from three manuscript review tools and revise their article based on these before it is posted on the platform as a preprint.
Penelope.ai can be used prior to submission to ensure the article complies with the platform’s ethical guidelines and policies. Running an article through this tool means authors are far less likely to miss key requirements and have their article returned to them at the very first hurdle. During the submission process, SciScore scans the methods for a variety of rigour criteria which have been shown to contribute to the reproducibility of research. It also detects elements such as sequencing data, plasmids, cell lines and software tools, and assesses how uniquely identifiable and accessible these are. These are elements that authors can easily include but may not have been trained in the importance of, and as such their work is less reproducible. Lastly, iThenticate scans the article for language, picking up on text that may have been unknowingly reused from previously published work. Our testing of this tool identified that a surprising number of authors do this, but it is easily rectifiable, and making these changes will improve the originality and scholarship of an author’s work.
A second unique advantage of our platform is the single location of all your article versions. Whilst many authors’ articles are posted on a preprint platform and subsequently published on a separate journal website, all preprint versions and the final published article will be together in one location on the Access Microbiology platform, with clear and easy links to navigate between them.
What do I need to know for the launch?
The funding from Wellcome Trust and the HHMI means that it will be completely free for authors to post and publish for the first 12 months after launch at the beginning of 2022, so we encourage early submission to take advantage of this. The sound science nature of the platform provides microbiologists with a place to publish all their work, as long as it is related to microbiology and the work is sound. We welcome any original research (including null, negative and replication studies), descriptions of large datasets, case reports, a new tool or code used to help researchers in their research, or even the description of a useful technique used in microbiology teaching or outreach. To read more on why sound science research is important and the types of research the Access Microbiology platform will publish, read our recent blog on Microbe Post.
We are living in a fast-paced and ever-changing period, and the Microbiology Society is embracing this with the Access Microbiology platform in the hope that we can provide a unique, transparent and trusted place for our community to publish their work.

Alex Howat
Open Research Platform Project Manager


