Microbiome-health associations: status and perspectives
09 May 2017
Over the last decade, there have been many exciting advances in the understanding of the microbiome – the community of organisms present in a particular location, often a sample from an animal, plant or the environment. Although research on the human microbiome grabs most of the headlines, many of the key methodologies were taken or adapted from environmental microbiologists who are used to dealing with very complex microbial communities that are very difficult to grow in the laboratory.
How we got here
We have known for a long time that humans and animals harbour a resident community of microbes on and in their bodies, but these micro-organisms are nutritionally fastidious and often strictly anaerobic, making their culture in the laboratory, identification and characterisation very challenging. The development of high-throughput DNA sequencing methods made it possible to apply culture-independent methods to construct inventories of organisms from any sample with sufficient numbers of cells to generate DNA for amplification by the polymerase chain reaction. Sequencing these pooled amplicons tells the investigator what bacteria are present. If there are even more cells to work with, the total pool of microbial DNA can be extracted and sequenced en masse (so-called ‘shotgun sequencing’), revealing not only what organisms are present, but also what genes they carry and what metabolism they are predicted to be capable of. Although the latter approach is much more expensive than simply profiling the sample, knowing their coding potential is usually much more biologically informative. Technical variations of this approach are used to identify mammalian viruses, bacterial viruses (bacteriophage), fungi, yeast and protozoa, but knowledge of these microbial communities in mammals is less developed than that of the bacterial microbiome.
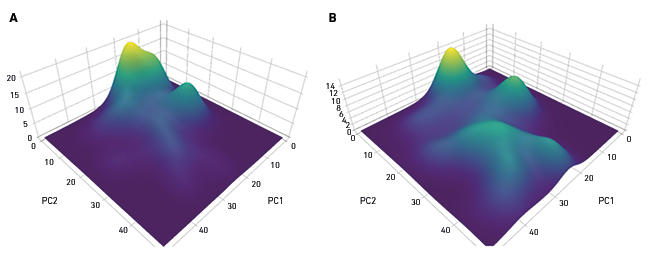
Studies of the human gut are currently more common than studies of other body sites, but the microbiome of skin, lung, oral cavity and liver, for example, are receiving increasing attention. A common denominator to many studies is the search for links between an altered microbiome composition and a disease or disease risk (Fig. 1). The list of such diseases linked to the intestinal microbiome includes obesity, inflammatory bowel disease, irritable bowel syndrome, type 2 diabetes and colorectal cancer, but associations of microbiome alterations with extra-intestinal conditions such as cardiovascular disease, and depression/behaviour are also being investigated. Because there are known ways or plausible new mechanisms whereby gut bacteria can influence inflammation, microbiome links to diverse conditions such as asthma, arthritis and frailty in older people are being investigated.
Fig. 1. Altered microbiome in health and disease. Principal coordinates analysis with density weighting of the microbiome composition in colonic mucosal biopsies of (A) healthy controls and (B) subjects with colorectal cancer.
The complexity of microbiome–host interactions
It has proven difficult to establish mechanistic links between microbiome alterations and disease, with one of the first conditions studied – obesity – proving most controversial of all. Rather than being able to fulfil Koch’s Postulates as one would for a bacterial pathogen, by finding the responsible organism and using a pure culture to replicate diseases symptoms in a model, it has not been possible to identify single organisms or definitive microbiome configurations that translate reproducibly from human to model systems and that cause obesity. This is probably due to a multiplicity of mechanisms by which the microbiome can influence metabolic health, involving diverse human processes such as calorie harvest from the diet, fat synthesis in the liver, inflammation, satiety, and levels and kinds of bile salts made in the liver to process fatty foods. Explaining how variations in a microbiome (composed of several hundred species, each with several thousand genes) interact with a multifactorial human pathophysiology was never going to be easy. Another high profile condition with microbiome links, inflammatory bowel disease (IBD), is apparently not characterised by a unifying set of depleted or over-represented micro-organisms across all patients and studies. Studies in twins suggest that up to half of the risk factors for IBD are environmental, so notwithstanding the accepted role of host genetic factors, it is highly likely that micro-organisms contribute to the disease in a manner as yet unclear.
Translating microbiome knowledge
The clearest role for the gut microbiome in disease is arguably Clostridium difficile-associated diarrhoea. Depletion of the gut microbiome by repeated antibiotic treatment allows a bloom of this organism (sometimes called C-diff) which can become recurrent and cause colitis and diarrhoea. A normal microbiome can be restored by faecal microbiota transplantation (FMT) in which the patient receives an enema or colonoscopic injection of a slurry prepared with the faeces of a healthy donor. The material is donated by a person selected by the recipient or their physician, or in some cases is acquired from a stool bank. FMT leads to 80%–90% cure rates in uncomplicated cases. The broadly accepted mechanism was simply the microbiome restoration, coupled with conversion by the incoming bacteria of primary bile acids to secondary and tertiary bile acids that inhibit C. difficile spore outgrowth. However, a recent small pilot study showed that sterile bacteriophage filtrates (not bacterial cells) also led to clinical remission, so even in this apparently ‘simple’ disease, the microbiome involvement may be more nuanced.
A characteristic of many diseases that feature microbiome alterations such as IBD, obesity, type 2 diabetes, and frailty in the elderly is loss of microbiota diversity, sometimes reflected in lower overall bacterial gene count. This is referred to by some authors as ‘dysbiosis’ but because two healthy people can have very different microbiota, dysbiosis is hard to reliably define. This makes routine microbiome profiling of healthy subjects of debatable value. Although identifying mechanisms linking microbiome alterations to health effects is still a work in progress, there is emerging evidence for the value of using microbiome markers to manage weight gain in obesity, predict post-prandial glucose in pre-diabetics, to stratify subjects with irritable bowel syndrome, and to detect colorectal cancer (see our recent review for details). Even more striking is the recent discovery that whether or not cancer patients derive clinical benefit from immunotherapy with checkpoint inhibitors (monoclonal antibodies that prevent inhibition of T-cell activation) is dependent on their baseline microbiota, which also determines whether or not they suffer from severe side effects that may require therapy withdrawal. These findings make profiling the gut microbiome not only clinically valuable (perhaps even sensible routine practice), but they also raise the prospect of rationally manipulating the microbiome to promote response to therapeutics in people who would otherwise be non-responders. A handful of drugs are already known to interact with the microbiome in ways that modulate their efficacy or side-effects, so this topic warrants considerably more attention.
Where to now?
When all the initial excitement of microbiome research dies down, we will realise that inventories of organisms and gene annotations are starting points, not end-points. There is a lot of microbiology research that needs to be done on microbial genes, enzymes, metabolites, cell models, pre-clinical models, and human subjects. Fastidious organisms will need to be cultured and grown as single cultures or as defined consortia, for example in artificial colon models (Fig. 2). Microbiologists will work intensively in their own labs, but also alongside bioinformaticians, metabolic modellers, cell biologists, physicians and epidemiologists, to unravel how the microbiome modifies risk for health and disease.

Fig. 2. Some aspects of the microbiome can be modelled by inoculating fermenter vessels with faecal samples, single isolated cultures or artificial consortia, and applying a range of analyses including transcriptomics and metabolomics to measure interactions and products.
Acknowledgements
Microbiome work in PWOT’s laboratory is supported by Science Foundation Ireland through a Centre award to the APC Microbiome Institute (SFI/12/RC/2273), and a Department of Agriculture, Food and the Marine (DAFM) award (11/F/053) for the ELDERFOOD project.
Dedicated to the memory of the late Professor George T. Macfarlane whose pioneering work on gut microbial ecology and biochemistry helped to lay the foundations.
Paul W. O’Toole
School of Microbiology & APC Microbiome Institute, Room 447 Food Science Building, University College Cork, T12 Y337 Cork, Ireland
[email protected]
Further reading
Lynch, S. V. & Pedersen, O. (2016). The Human Intestinal Microbiome in Health and Disease. N Engl J Med 375(24), 2369–2379.
O’Toole, P.W. & Flemer, B. (2016). Studying the Microbiome: “Omics” Made Accessible. Semin Liver Dis 36(4), 306–311.
Ott, S. J. & others (2016). Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium difficile Infection. Gastroenterology 152(4), 799–811.
Pitt, J. M. & others (2016). Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 44(6), 1255–1269.
Shankar, V. (2017). Gut microbiome profiling tests propelled by customer demand. Nat Biotechnol 35(1), 9.
Images: Coloured scanning electron micrograph of mixed oral bacteria. Steve Gschmeissner/Science Photo Library. Fig. 1. B. Flemer & P. W. O'Toole. Studying the microbiome: "Omics" made accessible. Semin Liver Dis 36(4), 306–311. Fig. 2. M. Perez & P. W. O'Toole.


