Candida albicans, a Dazzling Fungus: Application of a Novel Fluorescent Protein
My name is Jonas Devos, I’m working as a research assistant in the Laboratory for Molecular Cell Biology of Professor Patrick Van Dijck, KU Leuven, Belgium. This work started during my master’s thesis in the lab of Professor Van Dijck under the guidance of Dr Van Genechten and then continued after my graduation. The research lab of Professor Van Dijck mainly focuses on pathogenic fungi including Candida albicans, Candida glabrata, and Candida auris with topics ranging from nutrient sensing to the discovery of novel antifungal drug targets. Within these different topics, there is also a strong focus on the development of novel tools such as the fluorescent probe I present here.
In recent years fluorescent proteins (FPs) and fluorescence microscopy have become standard practice in molecular research. Since the original discovery of green fluorescent protein (GFP), a lot of different variants of fluorescent proteins have been made and discovered. The next generation of FPs are about half the size of GFP and require the interaction with a fluorogen to become fluorescently active. An example of such a protein is Fluorescence activating and Absorption Shifting Tag (FAST).
A classical FP, like GFP, can be compared to a lightbulb that can be turned on. Turning it off happens when the bulb breaks due to photobleaching (irreversible chemical changes) or degradation. The advantage of FAST over classical FPs is the fact that here the lightbulb, i.e. the fluorescence, is an exchangeable small molecule called the fluorogen. Addition of the fluorogen (the lightbulb) to the FAST protein (the bulb fixture) results in fluorescence. Furthermore, through addition and removal of the lightbulb, fluorescence can be turned on and off without breaking the protein. This allows spatial and temporal control of the fluorescence. It needs to be noted that FAST as a stand-alone protein is not fluorescent. A plethora of different fluorogens have been developed for FAST, each having their own spectral properties (each resulting in a different emission spectrum and thus colour).
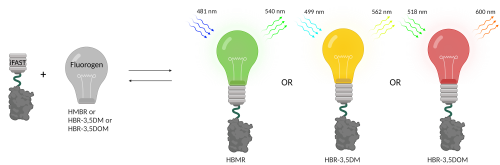
The (i)FAST system allows for great flexibility in both absorption and emission. In a live-imaging setting this one genomic tag can be used in different parts of the light spectrum to optimally fit other FPs or cellular staining. As the system is reversible, the fluorogens can be washed away and replaced with another.
Due to C. albicans being long overlooked as a significant pathogen, and because of its aberrant codon usage (it doesn’t follow the usual decoding system from DNA to protein), the palette of optimised fluorescent proteins for this organism is quite limited compared to the bacterial palette or the tags available for model organisms like Saccharomyces cerevisiae. In this work we present the use of a novel fluorescent protein, having multi-colour properties.
We show the application of an improved version of FAST (iFAST) in Candida albicans. Moreover, we show that the reversibility of the system can also be observed in the fungal species. Using the reversible FP system, we localized both an iron permease (Ftr1) shown to contribute to C. albicans virulence, and the main target of the azole class of antifungals, lanosterol 14α-demethylase (Erg11).
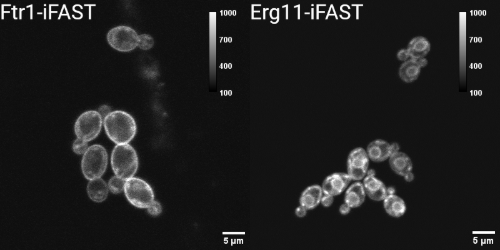
Within studying the reversibility and the localisation capacity of iFAST, we showed that both the integration of the system and the fluorogens have minimal influence on the growth of the cells.
During the research that went into this work, some variation between biological and technical repeats was observed. And thus, while there is still some room for improvement and optimalization, we believe that this publication can be an encouragement to optimize these novel tools for use in C. albicans or other species with a similar aberrant codon usage.
