Accessing the biosynthetic potential of Actinobacteria
Issue: Antimicrobials
01 November 2012 article

PAUL A. HOSKISSON
In an era of ever-increasing antibiotic resistance, can bacteria still provide a source of desperately needed novel antimicrobials?
The emergence of antibiotic-resistant micro-organisms was inevitable as soon as antibiotics were introduced to the vast array of drugs and treatments available to clinicians. Perhaps one surprise was the speed in which the first cases of resistance were observed in pathogens. Soon after, the race to find new antibiotics began in earnest. One source of potential new antibiotics was the Actinobacteria, one of the most abundant bacterial phyla on the planet, comprising up to 30% of the bacterial flora in some niches. Within their ranks are important human pathogens such as Mycobacterium tuberculosis, the causative agent of tuberculosis, but also the important antibiotic producers such as the Streptomyces species. Streptomyces is not the only genus that is useful industrially; indeed, many members of the phylum produce molecules that are useful as antibiotics, anticancer agents, antihelminthics or immunomodulators; these are often referred to as secondary metabolites as they are not essential for growth.

The discovery, by Selman Waksman, that the Actinobacteria were a good source of secondary metabolites, through the discovery of streptomycin production by Streptomyces griseus in 1943, meant that this group subsequently received an enormous amount of attention in order to add to our armoury of drugs. This arms race to find new weapons against resistant pathogens continued from the mid-1940s, through the golden age of antibiotic discovery in the 1960s, yet began to decline in the 1970s and 1980s. This decline in the introduction of new antibiotics was partly due to the rediscovery of the same molecules repeatedly in the same niches (Actinobacteria are common inhabitants of soil), and partly through the tightening of drug-licensing regulations, making it harder to bring new drugs to the market. However, with the need for new anti-infective drugs probably greater than ever, we need to be more cunning in our ability to discover and bring new antibiotics to the clinic.
THE POWER OF SEQUENCING
There are few, if any, areas of biology that have remained untouched by the power of whole-genome sequencing. The emergence of rapid, next-generation and third-generation sequencing technologies (454, Illumina, PacBio and Ion Torrent) has further advanced this progress at a pace, enabling rapid and cost-effective genome sequencing on the bench of any molecular biology laboratory. The first complete Streptomyces genome was published in 2002, through a collaboration between Professor Sir David Hopwood’s group at the John Innes Centre and the Wellcome Sanger Centre. Thereafter, the floodgates opened and there are over 100 Streptomyces genome projects listed currently on GOLD (www.genomesonline.org). One of the most surprising findings to emerge from all these genome sequences is the biosynthetic potential of these organisms. We have known for many years that most Streptomyces produce more than one secondary metabolite; however, studying the genome sequences has shown us that these organisms have the potential to produce many more, potentially useful metabolites. The model species, Streptomyces coelicolor A3(2), was known to produce three secondary metabolites – examining the genome sequence raised this number to over 20! This potential biosynthetic capability has been a recurring theme as each genome sequence is completed and the sequence-gazing begins. They are often referred to as silent gene clusters (the genes encoding secondary metabolites are often found together in clusters in the genome) as the products have yet to be ‘seen’ in cultures and are probably not expressed under the culture conditions that have been studied.
GIVING A VOICE TO THE SILENT MAJORITY
These previously unknown secondary metabolite gene clusters provide us with a challenge – to activate them and study the potential utility of their products. A range of approaches has been taken to study these clusters, including cloning them and expressing them in another Streptomyces species. This may enable the global regulatory processes controlling their expression in their native producer to be circumvented. Simple disruption of the pathway-specific regulators, which are easily identified within secondary metabolite clusters by bioinformatics, has been shown to result in activation of the genes within a cluster. Other approaches have included the manipulation of antibiotic genes through the use of short DNA sequences that interfere with regulatory proteins, thus ameliorating the effects of proteins that repress gene expression. The use of various stress and nutritional conditions has also yielded good results in terms of manipulating antibiotic production, such as the use of common soil carbon sources such as N-acetylglucosamine to activate expression of previously unobserved metabolites. This enhancing of secondary metabolism through the manipulation of cellular metabolism has great potential for awakening these silent clusters.
Recently there have been significant advances in the cloning of large DNA fragments directly from the environment. The gene clusters that encode secondary metabolites are generally in the region of 25–60 kbp in length; therefore, the ability to clone these genes from environmental samples is now a real possibility, often referred to as the metagenomic approach. This allows us to access novel secondary metabolites directly from the environment, without the need to culture the producing organism. These genes can then be expressed in a suitable heterologous host bacterial strain, of which there are many, including specially engineered strains of Streptomyces that don’t produce the majority of their own natural metabolites, and so are excellent producers of heterologous metabolites. One of the keys to identifying clusters using this approach is to explore previously neglected environmental niches, such as the deep-sea environment, hyper-arid deserts and Antarctica, all of which are yielding potentially interesting new antibiotics. The limitation of these approaches is the intensive screening required to identify biological activity in the cloned sequences.
SYNTHETIC BIOLOGY APPROACHES TO NEW ANTIBIOTICS
One of the most exciting approaches to emerge recently is the potential to apply synthetic biology to antibiotic production, discovery and manipulation. Synthetic biology can be thought of as the repurposing or redesign of biological systems for novel applications. The clustering of secondary metabolite genes offers a big advantage to the manipulation of their function, expression levels and production. Whilst synthetic biology is one of the current ‘buzz’ words in biology, this manipulation of antibiotic biosynthetic clusters is not particularly new. Since the mid-1980s, various groups interested in Streptomyces have been trying to make so-called ‘non-natural natural products’ using an approach called combinatorial biosynthesis (using genetic engineering to modify biosynthetic pathways to make new compounds). The ability to manipulate these clusters using molecular biology offers several advantages, not only in terms of generating novel activities, but also improving the pharmacokinetics of drugs, and reducing toxicity.
One group of secondary metabolites that has been amenable to combinatorial biosynthesis and has received lots of attention is the polyketides. Polyketides are synthesised by large multimodular enzymes that link simple precursors, such as acetyl-CoA or malonyl-CoA, in an iterative nature, growing chains that are tethered to an acyl carrier module on the enzyme. Manipulation of these multimodular enzymes then allows for alteration of the metabolite chain. Several methods have been used, such as those where simple gene disruptions can alter one of the chemical reactions during biosynthesis, resulting in longer or shorter chains, or without additional chemical decorations. Interestingly, one approach that has yielded amazing results is the formation of hybrid polyketides simply by transferring polyketide clusters into organisms producing related molecules. This allows the machinery from one cluster to modify the product in another in a very simple manner (see Fig. 1).
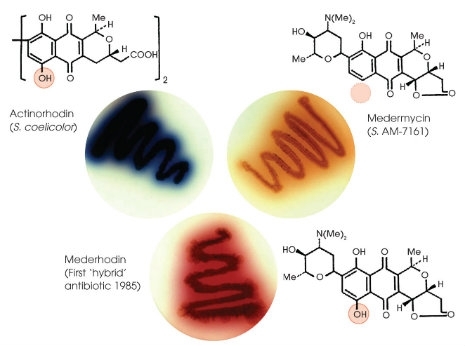
FIGURE 1
Hybrid polyketide biosynthesis. Streptomyces sp. AM-7161, making the brown-pigmented medermycin (top right) and S. coelicolor making the blue-pigmented actinorhodin (top left). The actinorhodin biosynthetic cluster was introduced into Streptomyces sp. AM-7161, creating the hybrid polyketide, which was called mederrhodin (bottom), which differed from medermycin in the addition of an -OH group. David Hopwood
TOWARDS A TULY SYNTHETIC BIOLOGY OF ANTIBIOTICS
The relatively simple experiments that have been carried out to date have wide-ranging implications; however, now we can couple these with the advances in molecular biology such as assembly of large DNA fragments, ligation-free cloning, and the advent of ‘BioBrick’ technology (www.biobricks.org) to create a truly synthetic biology for antibiotics. The multitude of secondary metabolite biosynthetic clusters available in the sequence databases means that we can assemble any enzyme simply from short oligonucleotides, introduce it into any biosynthetic cluster and examine the potential to alter the characteristics of the metabolite produced.
Of course, there are limitations to the modifications that certain chemical scaffolds are amenable to; however, we can envisage the very first, truly synthetic antibiotic biosynthetic gene cluster in the not-too-distant future. The assembly could be undertaken using BioBrick-type technology where each genetic unit is pieced together like LEGO bricks until a complete cluster is assembled. There is still a lot of work to undertake, as with any synthetic biology project. We need to understand and develop good regulatory sequences and promoters, and efficient ribosome-binding sites for manipulating expression levels and the overall logistics of piecing these together. Yet all of these approaches are tantalisingly within our grasp and will enable us to carry out bio-inspired chemistry synthetically in a bacterium in the near future.
ACKNOWLEDGEMENTS
Thanks to Professor Sir David Hopwood for the use of the picture in Fig. 1.
PAUL A. HOSKISSON
Strathclyde Institute of Pharmacy & Biomedical Sciences, University of Strathclyde, 161 Cathedral Street, Glasgow G4 0RE; Tel. +44 (0)141 548 2819; Email [email protected]
Image: Chiang, Y.-M., Chang, S.-L., Oakley, B.R. & Wang, C.C. (2011). Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol 15, 137–143. Medema, M.H., Breitling, R., Bovenberg, R. & Takano, E. (2010). Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat Rev Microbiol 9, 131–137. Winter, J.M., Behnken, S. & Hertweck, C. (2011). Genomics-inspired discovery of natural products. Curr Opin Chem Biol 15, 22–31..
