Comment: Pathogen genomics into practice: from promising research to real health impact
Issue: Fungal diseases
09 February 2016 article
Low-cost whole genome sequencing technology is catalysing a transformation in the way clinical and public health microbiologists and epidemiologists can manage the threat of infectious diseases. In comparison to existing microbiological methods, genomic methods offer greater sensitivity and specificity, as well as providing a description of a wide range of clinically and epidemiologically relevant characteristics of a pathogen, including identity, virulence determinants, drug resistance and relatedness to other pathogens.
In principle, genomics can be used to improve the management of infectious disease by allowing more precise diagnosis, detection and tracking of antimicrobial resistance and outbreak control. However, in practice evidence demonstrating that genomics improves patient care or population health, and is affordable when implemented in ‘real world’ pathways within the NHS, remains limited. Problematically, many of the benefits of genomics are expected to accrue only when it is deployed at scale, in an integrated health system-wide manner. As it is more or less impossible to pilot such an approach without significant capital and resource investment, a ‘calculated risk’ may have to be taken to commit to a genomic future for microbiology well in advance of all the necessary scientific evidence being available.
A nationally coordinated system of service development and delivery will be essential to realising the benefits of genomics for infectious disease management; the most fundamental components of this system being cross-organisational data integration, strategic coordination and strong leadership to manage the radical changes in microbiology service provision that will be required.
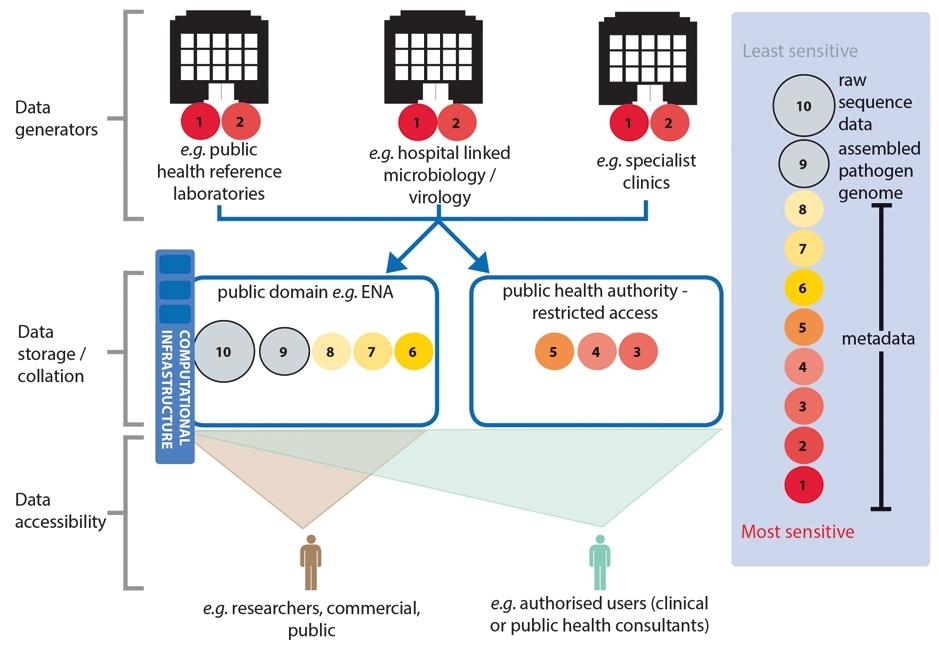
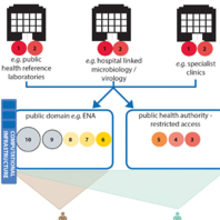
THE SIZE OF CIRCLES (NOT TO SCALE) ARE INDICATIVE OF THE RELATIVE DATA STORAGE BURDEN (COMPUTATIONAL DISC SPACE) OF THE DIFFERENT SUBSETS OF DATA. RAW GENOMIC DATA WILL CONSUME THE GREATEST DISC SPACE (THEREFORE COSTING MORE TO STORE THAN OTHER DATA TYPES) AND SO ITS LONGER TERM STORAGE WOULD BE BETTER SUITED IN A CONSOLIDATED REPOSITORY FOR HIGH-VOLUME DATA STORAGE.
Obtaining clinically useful information from pathogen genomes is reliant on our ability to compare them to other genomic data and combine this with relevant epidemiological or clinical information. The success or failure of genomics as a tool to improve infectious disease management will depend heavily, therefore, on the timely collation, integration and sharing of genomic and clinical/epidemiological metadata across the health system. These actions will also underpin the research and development needed to drive the expansion of the utility of genomics to a wider range of organisms and investigations than are currently feasible.
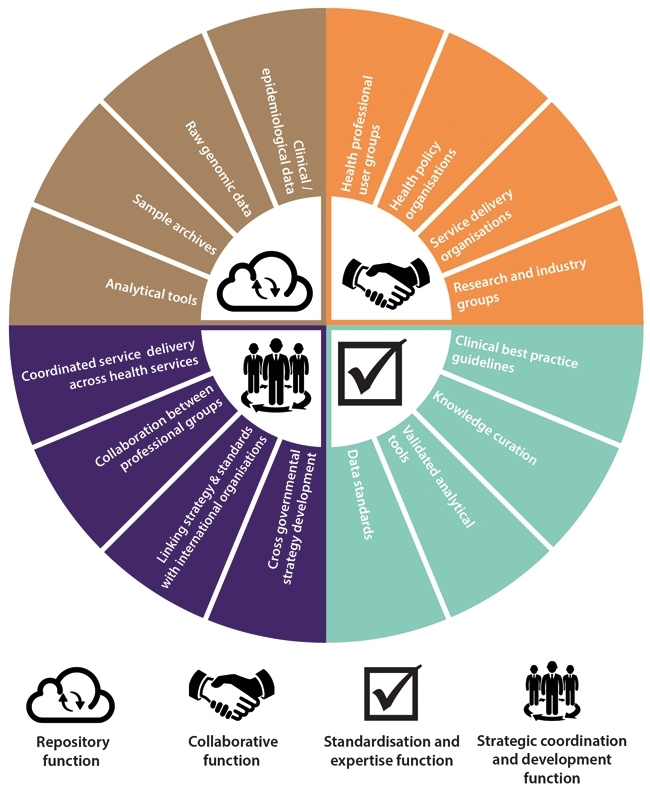
Achieving this vision of genomics-enabled microbiology services that are continuously innovating and expanding in scope requires the development of infrastructure to provide repositories for data, knowledge and samples, along with quality standards and data privacy safeguards to ensure their responsible use. This is no easy task considering the large number of different groups involved in producing and using this data, from front-line clinical staff, genomics researchers and clinical scientists, through to national agencies involved in food safety or animal health. The task is further complicated by the fact that infectious diseases do not recognise borders, and so international cooperation on these issues will also be essential. Although it may seem a daunting prospect, acting now to develop these mechanisms wherever possible, while many services are in the relatively early stages of development, will lead to future benefits as sequencing becomes more widespread and the need for a coordinated approach becomes ever more pressing.
Effective development of a national data management system to deliver the benefits of pathogen genomics now and in the future will not happen organically. Instead it will require strategic leadership and coordination. The various disciplines and organisations involved must agree on and work towards a common vision of how a future genomics-enhanced infectious disease management system will look. The benefits of such a cooperative approach are that the health system will be able to minimise duplication of effort, and share both resources and risks.
In addition to this ‘top down’ strategic collaboration, it will be equally important to bolster ‘bottom up’ cooperation amongst ‘front-line’ professionals. By sharing and developing knowledge, expertise and best practice around genomics, they can accelerate delivery of the highest quality care to patients and improve the protection of the health of our population. This means providing opportunities and systems to gather input from a wide range of professional groups, ranging from infectious disease physicians, medical microbiologists and infection control nurses, to clinical laboratory scientists and academic researchers, all of whom have a stake in realising the effective development and implementation of pathogen genomics services.
From our extensive analysis of the technology, health service, microbiology systems and extensive stakeholder consultation, we believe that genomics could, and indeed will be used as a front-line tool in the management of some infectious diseases in the near future. However, realising this vision necessitates overcoming current technological limitations and improving our understanding of the clinical and epidemiological significance of genomic variation, with the concomitant adoption of this new knowledge by health services in a timely manner. We are convinced this can be achieved if a prompt, concerted effort is made to share data, knowledge and expertise and if the strategic coordination and leadership needed to deliver a whole system approach are provided by those tasked with protecting the public’s health from infectious diseases.
SOWMIYA MOORTHIE AND LEILA LUHESHI
PHG Foundation, 2 Worts Causeway, Cambridge CB1 8RN, UK
[email protected]
FURTHER READING
PHG Foundation (2015). Pathogen Genomics Into Practice. www.phgfoundation.org/file/16849/. July 2015.
PHG Foundation (2015). Infectious disease genomics project. www.phgfoundation. org/project/id. Last accessed 7 December 2015.
Image: Both PHG Foundation..
