Adding dynamics to structure/function studies in microbiology: two-dimensional infrared spectroscopy
Issue: Future Tech
09 August 2016 article

Microbiology is a pioneering discipline. Many of the fundamental processes of biology were first elucidated in microbial systems – deciphering the genetic code, the one gene–one enzyme hypothesis, the mRNA hypothesis, the first genome sequences, and many more key biological breakthroughs were all the result of microbiologists! In an age where interdisciplinary projects are encouraged it is easy to be cynical. However, true collaborations between biologists and physicists can lead to fascinating insight into biological systems, especially when exciting new techniques can be applied to biology.
One of the central themes of biology is that structure is linked to function. At the molecular level, however, biology is dominated by intermolecular interactions, such as those between protein–ligand, protein–protein, protein–nucleic acid and nucleic acid–nucleic acid. As microbiologists, we often look for ways to characterise these kinds of interactions with a view to understanding our favourite (micro)biological system. These chemical interactions are underpinned by hydrogen bonding, and electrostatic or hydrophobic processes, occurring between the molecular partners. As we know, proteins are not static entities, and these intermolecular interactions are generally controlled by rapid structural fluctuations. For example, the typical lifetime of a hydrogen bond in solution is around 1 picosecond (to put this into context – 1 picosecond is to 1 second as 1 second is to 31,710 years). To fully understand and exploit the biology, therefore, it is necessary to complete the structure–function puzzle by incorporating dynamics. This kind of approach has proved very useful in terms of understanding protein–ligand binding, such as antibiotic molecules binding to their target proteins, or post-translational modifications of proteins.
In order to observe fast processes, you need a ‘camera’ with a fast shutter speed. One way to approach the understanding of protein dynamics with sub-picosecond temporal resolution is a relatively new experimental technique called ultrafast two-dimensional infrared spectroscopy (2D IR). 2D IR is uniquely sensitive to structural dynamics and it is capable of extending the range of information available to microbiologists studying molecular interactions in a complementary way to established tools, such as macromolecular crystallography, two-dimensional nuclear magnetic resonance (2D NMR), surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC). In terms of microbiological applications, 2D IR is a technique in its infancy and we are only now beginning to discover its potential. We feel 2D IR is capable of making a novel contribution to microbiology research areas, at a time when studies of protein–ligand interactions are fundamental to our understanding of globally relevant microbiological problems such as antimicrobial resistance and climate change.
Infrared (IR) spectroscopy is useful for studying proteins because the C=O bonds found in peptide linkages have stretching vibrational modes (so-called ‘amide-I modes’) that are intense and sensitive to the inter- and intra-molecular processes (think, school ‘mass on a spring’ type experiments) that underpin biological function, such as protein folding, ligand binding and enzymatic reactions. These are most commonly studied via IR absorption spectroscopy, which gives rise to a ‘one-dimensional’ plot of absorbance versus vibrational frequency (See Fig. 1a). However, the benefits of amide-I mode spectroscopy are restricted because the absorptions from individual amino acid residues in proteins overlap, or couple strongly to form delocalised vibrations that are due to protein secondary structure elements such as α-helices or β-sheets.
Multidimensional spectroscopy methods such as 2D IR measure the same molecular response as an IR absorption experiment, but spread the spectrum out over a 2nd frequency axis in order to uncover features that are not resolved in 1D. Practically, rather than simply measuring absorption, a 2D IR experiment consists of two IR laser pulses that hit the sample with a precisely controlled time delay between them. The first pulse excites the sample at a particular frequency (makes it vibrate) and the second laser pulse measures the effect of this on the molecule via an absorption spectrum. By progressively changing the wavelength of the laser for the first (pump) pulse, one builds up a 2D plot of excitation (pump) versus detection (probe) frequency. The time delay between the laser pulses allows us to examine a protein’s structure and its dynamics on the timescale of the pulse duration (~100 fs). This means that the IR absorption spectrum lies along the diagonal of the 2D IR plot (a surface response plot) while new information is found in the off-diagonal region of the spectrum (Fig. 1b). It is these off-diagonal regions that report on interactions between the structural components of our protein molecules – interactions between vibrations on specific amino acid residues or energy transfer processes within the molecule – essentially informing us on the structural interactions within proteins.
FIG. 1. (A) REPRESENTATION OF A 'ONE-DIMENSIONAL' IR SPECTRUM WITH TWO VIBRATIONAL MODES AND (B) 2D IR COUNTERPART TO THE SAME SPECTRUM, ILLUSTRATING COUPLING PATTERNS BETWEEN THE TWO VIBRATIONAL MODES. THESE TWO COUPLED VIBRATIONS WILL GENERATE OFF-DIAGONAL PATTERNS, AT COORDINATES INDICATED BY THE BLUE ARROWS.
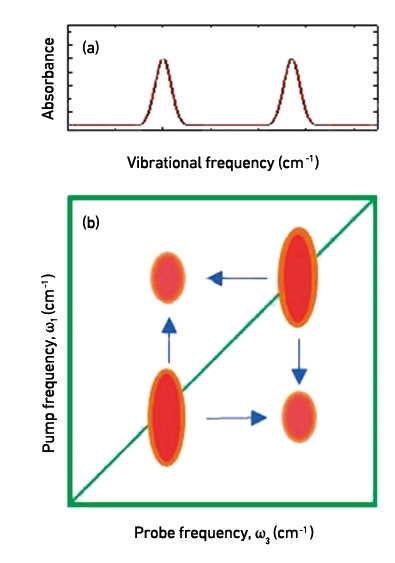
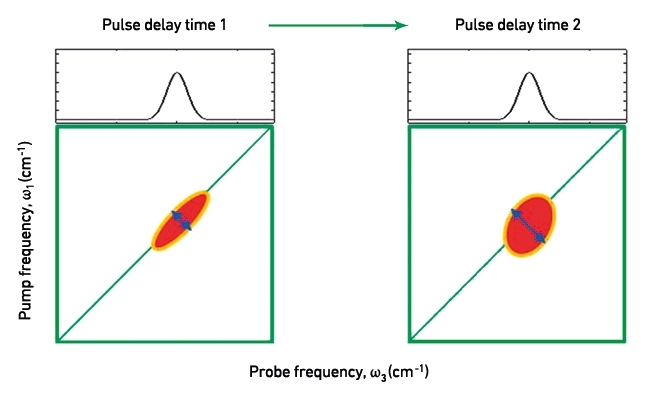
An example of a 2D IR spectrum is shown in Fig. 1(b). These can be thought of as a map of the vibrational modes within a molecule and how they interact. This off-diagonal region of the spectrum is richly structured, revealing patterns that are unique to secondary structural elements of the protein, but also reports on interactions between them. Most usefully, in terms of providing complementary information to established microbiological techniques, the anti-diagonal direction of the 2D IR spectrum reports on the flexibility of molecules, as structural motion allows vibrations to undergo small frequency changes during the time delay between laser pulses (Fig. 2). In turn, more flexible molecules give rise to greater anti-diagonal line broadening. These dynamics have been shown to vary sensitively with ligand binding and provide fundamental insight into this fundamental intermolecular contact. Thus, 2D IR provides a large quantity of new information but the difficulty lies in interpreting this in terms of biological function.
FIG. 2. 2D IR SPECTRA DEPICTING THE EFFECT OF MOLECULAR FLEXIBILITY ON THE ANTI-DIAGONAL WIDTH OF VIBRATIONAL MODES. WITH LONGER PULSE DELAY TIMES, IT BECOMES DECREASINGLY LIKELY TO FIND FLEXIBLE MOLECULES IN THE SAME POSITION OR ORIENTATION AS THAT AT THE FIRST PULSE. CORRESPONDINGLY, THEIR EXCITED VIBRATIONAL FREQUENCIES WILL ALSO EVOLVE WITH TIME, RESULTING IN ANTI-DIAGONAL LINE-BROADENING.
An important tool in this is to link 2D IR data to other methods in a complementary fashion. For example, the fast dynamics extracted by 2D IR bridge the gap between the atomic resolution of crystallography and the slower (µs–ms) dynamics accessible by NMR. Importantly, 2D IR measures protein dynamics on the exact timescale that computational simulations like molecular dynamics predict. These methods are used extensively to infer biological function, but until now experimental confirmation of the results has not been achievable.
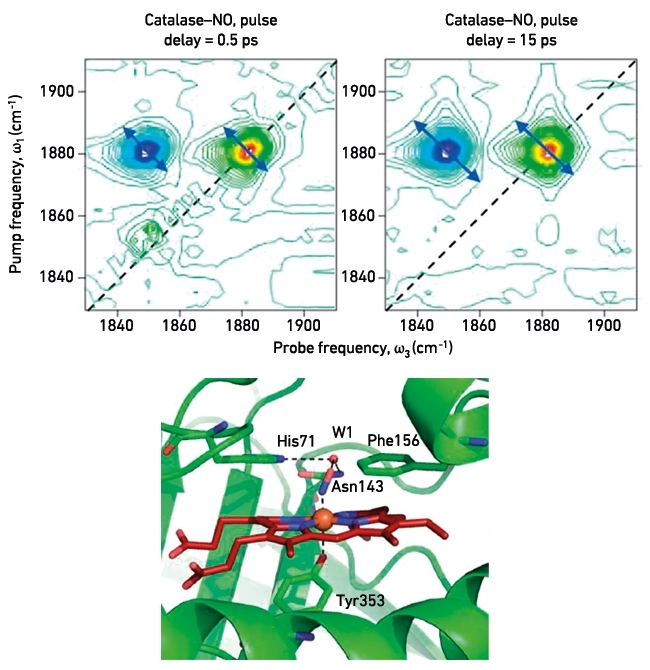
We have used 2D IR to study a range of systems, to shed light on the role dynamics plays in understanding microbiological processes. Recently, we applied 2D IR to an investigation of protein structural dynamics of the active site of catalase from the bacterium Corynebacterium glutamicum, and how this enzyme is inhibited by nitric oxide. By recording the dependence of anti-diagonal peak width of the NO group as a function of the time delay between laser pulses (Fig. 3a), we were able to show for the first time that mobile water molecules are present in the active site of the enzyme and that these are responsible for large-scale vibrational modes of the haem pocket that plays a key catalytic role in the enzymatic reaction.
FIG. 3. 2D IR DEPICTING THE ANTI-DIAGONAL LINE-BROADENING OF A NITRIC OXIDE MOLECULE BOUND AT THE ACTIVE SITE OF THE CATALASE. RAPID BROADENING OF THE NO VIBRATION INDICATES THE PRESENCE OF A NETWORK OF MOBILE WATER MOLECULES IN THE DISTAL CAVITY THAT ARE NOT RESOLVED IN THE CRYSTAL STRUCTURE, SHOWN BELOW.
Currently, we are using 2D IR to investigate antimicrobial drug resistance, specifically the inhibition of the essential enoyl reductase (InhA) from Mycobacterium tuberculosis by the frontline anti-TB drug isoniazid. We have studied the components of the active isoniazid-NADH complex and are now combining standard molecular biology, such as site-directed mutagenesis and enzymology to augment the 2D IR studies and develop a clear picture of the active site structure–function–dynamic relationship. We have been able to recreate mutations within the InhA protein that lead to clinical resistance and study how these behave when exposed to isoniazid and also natural enzymatic substrates. Understanding the role of molecular dynamics in relation to structure and function can aid the design of novel anti-TB drugs in the future and will hopefully lead to the recognition that 2D IR has a key role to play in microbiology.
Currently, 2D IR is not a bench-top experiment, with each spectrometer being built by the users. However, this was the situation that NMR was in 40 years ago and now it is a standard experimental technique. 2D IR has great potential for microbiology and biochemistry; there are many problems in microbiology that can be assisted with such techniques – so be open to collaboration and befriend a tame physicist!
NIALL SIMPSON, NEIL T. HUNT
Department of Physics, SUPA, University of Strathclyde, 107 Rottenrow East, Glasgow G4 0NG, UK
PAUL A. HOSKISSON
Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, 161 Cathedral Street, Glasgow G4 0RE, UK
[email protected]
@paulhoskisson
FURTHER READING
Adamczyk, K. & others (2012). Measuring protein dynamics with ultrafast two-dimensional infrared spectroscopy. Meas Sci Technol 23, 062001.
Adamczyk, K. & others (2015). Ultrafast infrared spectroscopy reveals water-mediated coherent dynamics in an enzyme active site. Chem Sci 6, 505–516.
Bagchi, S. & others (2010). Conformational switching between protein substates studied with 2D IR vibrational echo spectroscopy and molecular dynamics simulations. J Phys Chem B 114, 17187–17193.
Hunt, N. T. (2009). 2D-IR spectroscopy: ultrafast insights into biomolecule structure and function. Chem Soc Rev 38, 1837–1848.
Ishikawa, H. & others (2007). Neuroglobin dynamics observed with ultrafast 2D-IR vibrational echo spectroscopy. Proc Natl Acad Sci U S A 104, 16116–16121.
Shaw, D. J. & others (2015). Multidimensional infrared spectroscopy reveals the vibrational and solvation dynamics of isoniazid. J Chem Phys 142, 212401–212409.
Simpson, N. & Hunt, N. T. (2015). Ultrafast 2D-IR spectroscopy of haemoproteins. Int Rev Phys Chem 34, 361–383.
Simpson, N. & others (2013). Infrared spectroscopy of nicotinamide adenine dinucleotides in one and two dimensions. J Phys Chem B 117, 16468–16478.
Thielges, M. C. & others (2011). Protein dynamics in cytochrome P450 molecular recognition and substrate specificity using 2D IR vibrational echo spectroscopy. J Am Chem Soc 133, 3995–4004.
Image: Dr Neil Hunt operating a 2D IR spectrometer at the University of Strathclyde. Tim Briggs. Figs. 1, 2 and 3. Niall Simpson..
