An (in)sensitive pathogen – Mycobacterium tuberculosis
Issue: Mycobacteria
27 August 2014 article
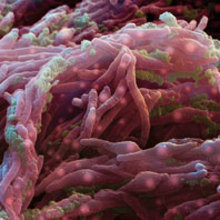
Mycobacterium tuberculosis is one of the principal human pathogens on the planet. It infects around 2 billion people worldwide annually, causing almost 1.3 million deaths in 2012.
Most infected individuals do not go on to develop an active disease, but will remain latently infected throughout their lifetime. Nonetheless, 5–10% of infected individuals will develop the life-threatening form of tuberculosis, with symptoms such as fatigue, weight loss, enhanced sweating and coughing, and even coughing up blood. If it is untreated or undiagnosed until its late stages, tuberculosis may lead to severe lung damage and death.
What does Mycobacterium tuberculosis look like?
Not only is tuberculosis one of the most common bacterial infections that causes death to humans, but it is also one of the oldest known to humanity. Tuberculosis was recently found to be the most probable cause of death of several Egyptian mummies from thousands of years ago. However, it was only at the end of the 19th century when Robert Koch first isolated M. tuberculosis and postulated that this was the ‘evil’ causing tuberculosis. Microscopic observations revealed that the bacterium is rod-shaped, 2–4µm in length and 0.2–0.5µm in width.
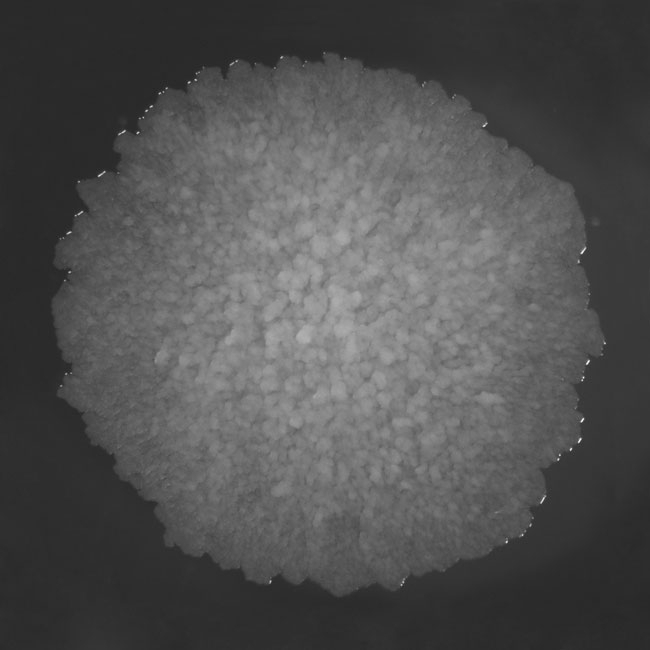
It only grows on rich media and divides every 16–20 hours, which is very slow compared to other bacteria, such as Escherichia coli, which can multiply every 20 minutes. M. tuberculosis requires oxygen to grow and forms rough, wrinkled colonies on solid culturing media.
Since the genome sequence of the M. tuberculosis H37Rv laboratory strain was determined in 1998, a new door for scientists has been opened and this has produced a better understanding of the biology of this pathogen. First of all it became apparent that the genome of M. tuberculosis is rich in G+C nucleotides (~70%) and it contains information for about 4,000 open reading frames, encoding many proteins involved in the cellular process of lipogenesis and lipolysis.
What makes M. tuberculosis a worldwide pathogen?
The success of M. tuberculosis as a pathogen is mainly due to its insensitivity to most known antibiotics and its ability to precisely sense the immune host responses and adequately adapt to their lifecycle.
Mycobacterial resistance to antibiotics is mostly due to the extraordinary composition of their cell wall. It is truly a bacterial fortress, with a thick and complex multilayer cell envelope that is impermeable to numerous compounds. The outmost layer of the mycobacterial coat is called the capsule, and is formed mostly of glycans that are directly exposed to cells of the human immune system, primarily macrophages, and they provide an antiphagocytic shield. Underneath the capsule there is a layer of lipids called mycolic acids, decorated with porins and attached to yet another layer of sugary arabinogalactan. Arabinogalactan is in turn attached to the peptidoglycan and both structures are intercalated with lipomannan and lipoarabinomannan.
FIG. 1. THE COLONY MORPHOLOGY OF M. TUBERCULOSIS GROWN ON SOLID MEDIA
The complex cell-wall composition together with efflux mechanisms and a battery of antibiotic-degrading enzymes account for natural resistance of mycobacteria against β-lactams, tetracycline, fluoroquinolones, aminoglycosides and many other chemotherapeutics. Due to the low effectiveness of practically any single antibiotic therapy that would be safe for humans, the treatment of tuberculosis usually takes 6–12 months and combines four first-line anti-tuberculosis drugs: isoniazid, rifampin, pyrazinamide and ethambutol or streptomycin. This expensive therapy must be taken under a strict regime to avoid the development of bacterial resistance. If the first-line drugs turn out to be ineffective, a combined therapy of second-line drugs must be considered (amikacin, kanamycin, capreomycin). These drugs are generally more expensive and/or have more pronounced side effects and are, thus, less safe to use in humans.
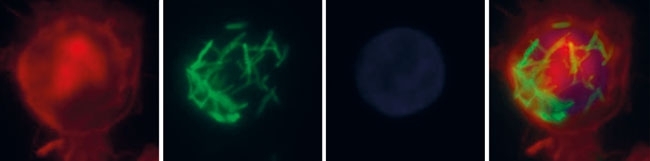
FIG. 2. PHAGOCYTOSIS OF GFP (GRESCENT PROTEIN)-TAGGED M. TUBERCULOSIS H37RV BACTERIA BY U937 CELLS, ENTIATED TO MACROPHAGES WITH POKWEED MITOGEN (PMA)
M. tuberculosis is not only resistant to the bactericidal effects of many antibiotics, but it can cope well with the naturally occurring mechanisms of human immunity responses. It can shift between an actively replicating, highly infectious state and a physiologically muted – dormant – state, according to the host’s immunity status. The phenomenon of dormancy is believed to be regulated via the two-component regulatory system DosR and DosS (or, alternatively, DosT), one of the sensing machineries of the bacterium. The tubercle bacilli contains several two-component regulatory systems, most of them paired, of a sensor histidine kinase and a response regulator. Sensor histidine kinases are usually membrane-bound proteins that, upon detecting an environmental signal, undergo autophosphorylation transferring the phosphoryl group onto the regulatory domain of the cytosolic response regulator. This in turn activates the output domain of the response regulator, which, in most cases, possesses DNA-binding properties and promotes or represses the transcription of selected genes. The canonical two-component systems in mycobacterial models were studied extensively in recent years. M. tuberculosis possesses 11 such two-component systems, e.g. SenX3/RegX3, PhoP/PhoR, DosR/DosS and MtrA/MtrB. Out of the known genetically linked regulatory systems, only PrrA/PrrB and MtrA/MtrB systems are believed to be essential for in vitro growth as well as for virulence of M. tuberculosis. Interestingly, the genome of M. tuberculosis possesses information for some orphaned two-component regulatory system elements, namely six response regulators and three sensor histidine kinases. The role of such orphaned elements in the bacterial response to environmental changes is fragmentary and remains to be determined. However, it is believed that such orphaned elements are involved in regulation of virulence and pathogenicity processes.
Development of drugs
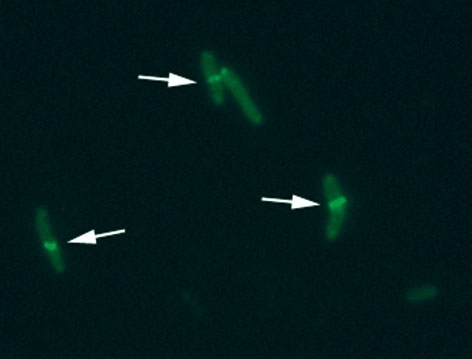
In recent years, observations of the increasing number of multidrug-resistant or even totally drug-resistant strains of M. tuberculosis, as well as co-pandemics with HIV-linked tuberculosis, has emphasised the need for the discovery of a new generation of antimicrobial agents. However, better understanding of the biology of the bacterium itself is fundamental for developing new and effective strategies to combat tuberculosis. Multiple studies are currently carried out worldwide, trying to identify new potential therapy targets. The characteristics of possible targets include protein elements that are involved in multiplication of the bacteria and other crucial processes like cell wall biosynthesis, DNA replication and repair, transcription, translation or virtually any protein that carries out an enzymatic reaction that is essential for bacterial survival. Inhibition of the activity of such protein candidates must not cause interference with human physiology. Ideally, chemotherapy that would interfere with the bacterial cell division or cell cycle mechanisms could prevent the pathogen from proliferating and stop the infection. Unfortunately, mycobacteria differ significantly from other bacteria in some aspects of their biology. For instance, the cell division process in M. tuberculosis is poorly understood so far. However, as in other bacteria, FtsZ, the prokaryotic homologue of eukaryotic tubulin, appears to play a prominent role in the cytokinesis of this organism. FtsZ assembly into a circumferential ring at the mid-cell position initiates recruitment of several other proteins involved in this process and enables formation of the division septa. Some scientists consider FtsZ protein as a very promising target for antimicrobial drug discovery, primarily due to its leading and irreplaceable role in the bacterial cell division process, and, secondly, due to its well-studied structure and biochemical activity.
FIG. 3. FUSION OF FTSZ WITH GFP IN ORDER TO VISUALISE Z-RING (INDICATED BY ARROWS) FORMATION IN M. TUBERCULOSIS CELLS
A list of other proteins being considered as potential candidates for new drug targets are NAD+-dependent DNA ligase A and primase DnaG, both essential for growth and survival of M. tuberculosis cells and, importantly, present only in bacteria and not in humans. Another group of interesting targets include proteins involved in the biosynthesis of mycolic acids and arabinogalactan and the biosynthesis of isoprenoids; however, these are mostly exclusive to mycobacteria and are less likely to be active against any other bacteria.
Currently, there are numerous ongoing and developing studies trying to develop therapeutic agents that will help to efficiently eradicate M. tuberculosis from the systems of infected individuals. With continued financial support from various organisations and a joint effort from the scientific community to study tubercle bacilli, such developments will hopefully happen in the near future.
ACKNOWLEDGMENTS
Work was supported by grant: Iuventus Plus IP2011 042571.
RENATA PLOCINSKA, JAKUB PAWELCZYK & JAROSLAW DZIADEK
Institute of Medical Biology, Polish Academy of Sciences, Lodowa 106, 93-232 Lodz, Poland
[email protected]
FURTHER READING
Bretl, D. J. & others (2011). Adaptation to environmental stimuli within the host: two component signal transduction systems of Mycobacterium tuberculosis. Microbiol Mol Biol Rev 75, 566–582.
Centers for Disease Control and Prevention (2006). Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs – worldwide, 2000–2004. Morb Mortal Wkly Rep 55, 301–305.
Cole, S.T. & others (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544.
Dziadek, J. & others (2002). Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial host. Microbiology 148, 961–971.
Errington, J. & others (2003). Cytokinesis in Bacteria. Microbiol Mol Biol Rev 67, 52–65.
Gorna, A. & others (2010). DNA repair systems and the pathogenesis of Mycobacterium tuberculosis: varying activities at different stages of infection. Clin Sci (Lond) 119, 187–202.
Image: Scanning electron micrograph of M. tuberculosis. Eye of Science/SPL. Fig .1. Jakub Pawelczyk. Fig. 2. Marcin Bartlomiejczyk. Fig. 3. Renata Plocinska..
