Comment: Badgers and bovine TB - how can environmental microbiology help?
Issue: Mycobacteria
27 August 2014 article
It is well recognised that the incidence of bovine tuberculosis (bTB) in the UK has been growing alarmingly in recent decades. The problem of bTB in cattle has affected a number of countries worldwide – notably Ireland, USA, New Zealand and many countries in Africa.
In all cases a reservoir of the disease has been identified in wildlife ranging from badgers in the UK and Ireland, to possums in New Zealand and white-tailed deer in the USA. However, the mode of transmission between wildlife and cattle is the subject of debate and once the disease becomes endemic the likelihood of cattle-to-cattle transmissions complicates the routes of transfer. Several years ago I became interested in the environmental aspect of this problem following discussions with a fellow microbiologist, Eamonn Gormley, who was working on bTB at the Veterinary Science Centre at University College Dublin (UCD). We developed an idea to investigate environmental contamination of pasture and soil surrounding setts where badgers lived, and determine if we could detect the causal agent Mycobacterium bovis. Working with Orin Courtenay, an epidemiologist at the University of Warwick, we considered whether faecal shedding of the bacterium could be a useful measure of transmission risk from badgers.
Previous studies have shown that cultivation of M. bovis from badger faeces lacked sensitivity due to contamination from the wide range of other bacteria in the samples and low numbers of cells (positive culture from 27/1064 samples of faeces). In addition, cultivation of a pathogen outside of its host is likely to reduce culturable status due to stress and the normal practice of decontamination with strong alkali proving too harsh when cells were recovered from soil. This led us to develop molecular assays for M. bovis in faeces and these proved useful to investigate shedding status of social groups. It should be noted that faecal shedding is an indication of infectiousness. Our studies with M. bovis also proved that polymerase chain reaction (PCR) detection in soil and faeces was a reliable indicator of intact cells as the turnover of cell-free DNA in soil and other substrates was rapid.
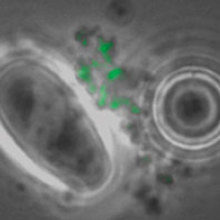
FIG. 1. M. BOVIS CELLS CAPTURED BY MAGNETIC BEADS WITH FITC M. BOVIS MONOCLONAL ANTIBODY, WITH DAPI STAIN AND IMAGE OVERLAY WHITE LIGHT AND FITC. A LARGE OVAL PROTOZOAN IS VISIBLE IN ALL IMAGES, EXCEPT FITC. BARS, 3ΜM
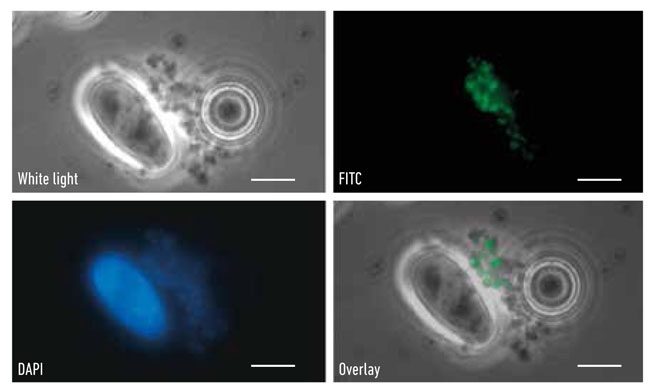
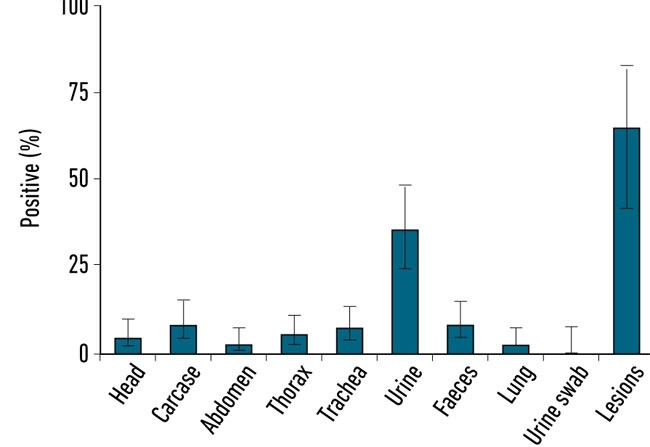
During the past decade, molecular detection of pathogens has become an accepted method of diagnosis, especially where antibiotic resistance was an important attribute as in the case of methicillin-resistant Staphylococcus aureus (MRSA) in blood and multidrug-resistant (MDR) Mycobacterium tuberculosis in sputum. Sampling wildlife is demanding and usually requires either live capture or culling to determine disease status but the collection of badger faeces is non-invasive and does not involve any disturbance of the sett. So the possibility of using molecular detection of M. bovis in faeces was investigated. The badgers dig latrines near their setts facilitating the collection of faeces and samples to be examined by microscopy to identify the presence of M. bovis cells; these can be visualised using immunofluorescence staining (Fig. 1) and quantified in the lab by quantitative (q)PCR. In collaboration with Eamonn Gormley, we were able to compare PCR positives in tissue from diseased badgers and levels in tracheal swabs were comparable to rectal faeces across a range of sampled individuals (Fig. 2). Shedding (trachea, urine and faeces routes) correlated with animals exhibiting more severe disease status. In order to rigorously test and validate the qPCR assay (RD4scar) a number of studies were done in collaboration with the Animal Health and Veterinary Laboratories Agency (AHVLA) Weybridge and Woodchester Park providing opportunities for ring trials and comparison with results from long-term badger monitoring.
FIG. 2. THE DETECTION OF M. BOVIS VIA QPCR RD4SCAR ASSAY IN BADGER TISSUES
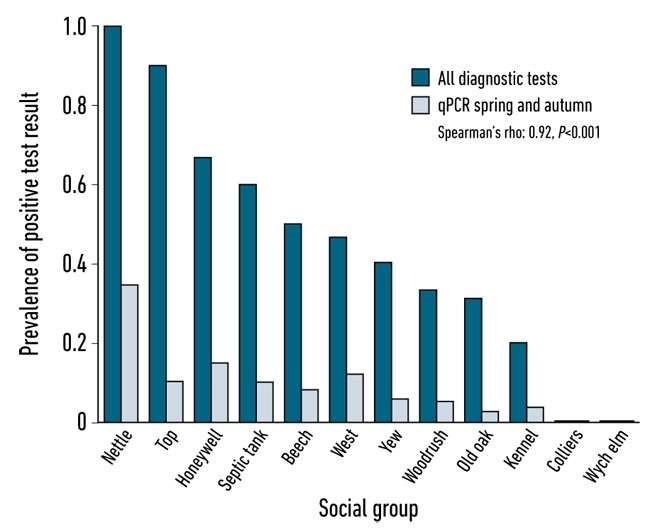
The latest study involved direct comparison of qPCR data with badger disease status obtained via trapping and the application of diagnostic tests. Trapping and testing of the badgers associated with the social groups under study was carried out by AHVLA Woodchester Park using IFN-gamma, Stat-Pak and culture of clinical samples to determine social group level disease prevalence. The faecal latrine sampling across the whole year was performed in close conjunction with the badger trapping and testing programme allowing comparison with the individual diagnostic tests as well as group bTB status. qPCR results varied by season, with spring and autumn exhibiting 100 and 80% sensitivity, respectively, against the combined trapped badger diagnostics for the same season. The degree of infection within a social group (trapped badger diagnostic test results) was strongly correlated with the degree of shedding as determined by faecal qPCR (Spearman’s rho=0.92, P<0.001) (Fig. 3). Bacterial loads varied considerably between social groups. We suggest that a small number of high prevalence social groups may be responsible for the majority of M. bovis that is shed into the environment and therefore present the highest potential risk of onward transmission.
FIG. 3. CORRELATION BETWEEN A SUITE OF DIAGNOSTIC TEST RESULTS CARRIED OUT ON TRAPPED BADGERS SHORTLY BEFORE FAECAL SAMPLING IN SPRING AND AUTUMN AT BADGER SOCIAL GROUP LATRINES IN WOODCHESTER PARK
Any effective disease control strategy needs reliable monitoring of both infection status as well as individual or group transmission risk, to maximise cost-effective control measures such as vaccination and improved biosecurity. In our view, understanding transmission pathways by monitoring the pathogen non-invasively in environmentally shed samples is an important component of the solution to the current bTB crisis.
ACKNOWLEDGEMENTS
The authors are grateful for funding from the Department of Environment, Food and Rural Affairs (Defra) and the Biotechnology and Biological Sciences Research Council (BBSRC). We wish to thank AHVLA scientists Glyn Hewinson, Jason Sawyer, Dez Delahay, and UCD scientists Eamonn Gormley and Leigh Corner, and their respective research teams for collaboration. We thank our research team for their excellent field and lab work.
ELIZABETH M. H. WELLINGTON & ORIN COURTENAY
School of Life Sciences, University of Warwick, CV4 7AL
[email protected]
FURTHER READING
Courtenay, O. & others (2006). Is Mycobacterium bovis in the environment important for the persistence of bovine tuberculosis? Biol Lett 2, 460.
Travis, E. R. & others (2011). An inter-laboratory validation of a real time PCR assay to measure host excretion of bacterial pathogens, particularly of Mycobacterium bovis. PloS One 6, e27369.
Wilesmith, J. W. & others (1986). Tuberculosis in East Sussex. IV. A systematic examination of wild mammals other than badgers for tuberculosis. J Hyg 97, 11.
Young, J. S., Gormley, E. & Wellington, E. M. (2005). Molecular detection of Mycobacterium bovis and Mycobacterium bovis BCG (Pasteur) in soil. Appl Environ Microbiol 71,1946.
Watch Professor Wellington's Hot Topic lecture at the Society's 2014 Annual Conference on bovine TB.
Image: Fig.s 1, 2, 3. E. M. H. Wellington..
