The microbial world at our feet
Issue: Soil
07 May 2015 article

A handful of soil may contain greater than a billion micro-organisms, and the total weight of bacteria, archaea and fungi in your back garden is measured in kilograms and contains at least 10,000 different genotypes. If you extracted the microbial DNA from just one gram of soil and stretched it out it would extend more than 1,500 kilometres!
Nowadays, we tend to talk not about microbial species but operational taxonomic units (OTUs). This is because horizontal transfer of DNA between bacteria housed within biofilms at organic matter and plant root surfaces, coupled with mutation and selection pressures, are commonplace. In other words, the soil microbiota is dynamic and evolving.
So many microbes and so much to do
Some soil microbes are competing or collaborating in their attempts to capture the limited resources and occupy the prime real estate. Others lay dormant or are just ticking over waiting for the environment to shift from dry to wet, aerobic to anaerobic, cool to warm, for the arrival of a plant root, the death of an adjacent microbe, or the appearance of an exotic substrate that the majority of the population doesn’t recognise.
The soil is a massive and versatile repository of activities capable of responding to any number of chemical, physical and biological conditions. The microbes are the agents for many of the processes that keep our world alive: carbon and nitrogen cycling, generating essential plant nutrients, degrading potentially dangerous chemicals, controlling plant pathogens, building and retaining soil structure, forming symbioses with plants, sequestering carbon, and so on. So, can we be complacent and assume that microbes will continue to do their job regardless of how we treat them? This is an important question and must be framed in the context of climate change with its associated increases in temperature and carbon dioxide, more frequent extremes of weather conditions, and the consequential impacts on the above-ground flora and fauna.
Persuading microbes to help us
Exploiting microbial activities has been an ambition of soil biologists for more than a century, yet the successes with legumes and nitrogen-fixing bacteria and plants and phosphorus-scavenging mycorrhizal fungi have been slow to expand to include other activities. Microbial biocontrol and bioremediation have been two of the ambitions with more disappointments than successes.
One of the problems is that we have been poor at growing soil microbes in the laboratory. Microbiologists have been slow to develop recipes for media and to understand the complex environmental factors needed for in vitro reproduction. There may be more than 50,000 bacteria, archaea and fungi isolated, cultivated and described but these represent less than 5% of the total soil population.
Developments in microscopy, modelling and, above all else, molecular biology are providing us with information about these difficult to grow microbes. At the heart of these advances is the ‘omics’ revolution: genomics, proteomics, transcriptomics and metabolomics. ‘Terragenomics’ are here to stay.
The new techniques are telling us more about the potential of microbes and how, why, when and what they express in the ever-changing soil microenvironment. Of course, the astronomical volume of data generated demands substantial computational resources if they are to reveal their secrets. But even with sophisticated equipment and limitless computer access, sequencing the entire soil microbial genome is a daunting challenge. And that is even before we try to understand how each strain works independently and collectively. It may be that treating the soil as a complex multicellular organism rather than individual OTUs will enable us to set more realistic and achievable targets.
Plants and microbes talking among themselves
Rhizosphere numbers may be many times more than those in the root-free soil as microbes migrate to the root surface in response to the dozens of signals and substrates exuded by the plant. The most integrated of plant–microbe relationships (mycorrhizas) involve fungi and more than 90% of land plants and may represent the earliest examples (~450 million years ago) of the co-evolution of plants and microbes in soil. The other major integrated plant–microbe symbiosis, involving legumes and rhizobia, is closely linked to mycorrhizal activities. The fungi and the bacteria work together with the plant for their mutual benefit even though the tripartite relationship may be fragile and a trade-off under the prevailing circumstances.
In recent years we have identified more and more of the signals that microbes and plants use to communicate. Many involve acyl homoserine lactones (AHLs), flavonoids and peptides in what is called quorum sensing but any potential substrate may act as a chemo-attractant as long as it sets up a concentration gradient in the aqueous phase. Plants not only engage in this mutually beneficial chatter but also fight back against any negative or competitive challenges. There are AHL mimics that fool small numbers of potential phytopathogens into synthesising and secreting invasive enzymes long before there are enough of them to do any damage. Then there are compounds that block microbial receptors so that the signals they send to each other are not detected.
Microbes talking to other microbes trigger individual and collective responses in order to exploit potential substrates. This type of cooperation is essential in the breakdown and metabolism of complex macromolecules, such as lignocellulose: a process that involves dozens of OTUs and many different extracellular and intracellular enzymes.
Getting rid of the mess
Many soils suffer from chronic or acute pollution from industrial sources and are rendered unsuitable for human habitation, agriculture and recreation. In addition, contaminated soil acts as a source of organic and inorganic toxins for water, the atmosphere and wildlife in general. There are millions of hectares that are in need of remediation and, for a long time, exploiting the catabolic potential of the microbial world has had great appeal. An optimistic view might be that microbes are so smart that they will break down all pollutants and render them harmless. All we need to do is to persuade them to set about saving our soil.
Sometimes chronic pollution serves as a selection pressure and competent degraders are already present in the soil. But other factors may limit the expression of their degradative potential: pH, compaction and poor oxygen penetration as well as electron acceptor/donor concentrations and carbon-to-nitrogen ratios. Some of these constraints can be overcome by in situ biostimulation (e.g. rotovation, liming, adding organic matter). If this doesn’t work, bioaugmentation with a cocktail of microbes may be the remedy.
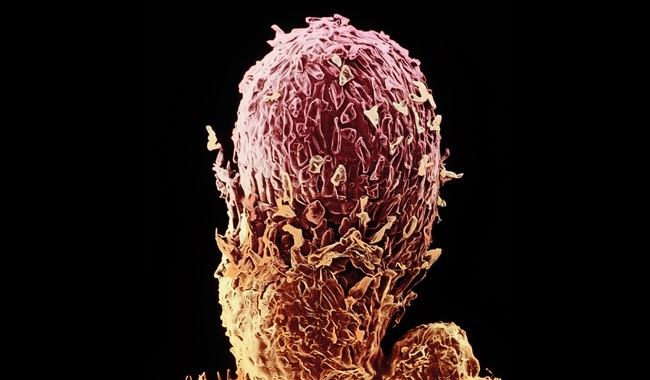
Large numbers of bacteria and fungi have been generated in laboratories using the target pollutant(s) to select competent microbial populations. The majority of these isolates fail when applied to soil. The reasons for this are not hard to find. If you were to name all the properties of a successful bioremediation inoculant the list would go on for some time. Catabolic potency – yes, but this needs to be expressed at an appropriate level by the bacteria and fungi, which must stay focused on the xenobiotic and not switch their attention to other more easily available and higher energy yielding natural compounds. The inoculant also needs to be a good competitor in an established and aggressive microbial community, detect and migrate to its unevenly distributed target, have affinity for the differing concentrations of the pollutant, and be physiologically versatile by functioning at various temperatures, redox conditions and pH values. The best biodegrader might even need to form a close partnership with indigenous microbes or plants to achieve the ultimate goal of converting the pollutant into carbon dioxide and water.
Another constraint to successful bioremediation is that the pollutants may be bio-unavailable because they are bound to clays and humic matter, hidden within soil aggregates, or hydrophobic and much too large (e.g. many aromatic hydrocarbons) to enter the microbial cell without prior disintegration. In these cases, ‘remediationists’ have turned to surfactants to increase the solubility of the troublesome compounds and considered using depolymerases to generate small molecular mass products that the cell can recognise. Knowledge of the catabolic stages involved in a pollutant breakdown may reveal a small number of rate-limiting steps. In this context, genetically manipulating indigenous microbes to express dehalogenases is a promising avenue for research.
As important as pollutant breakdown is, the rate at which decontamination takes place, tracking the inoculant and monitoring the products are essential. Reporter genes, introduced into the biodegraders or other soil microbes, can detect end-points and prescribed targets as well as informing risk assessment, and are necessary components of any credible bioremediation protocol. Too many snake oil remedies have already hit the market.
There are more than 250,000 polluted sites in the EU alone with a remediated value of twice the gross annual product. These are the numbers that influence politicians and funding agencies. Bioremediation must play a major role in our stewardship of soil in the 21st century.
Soil microbes are as important as plants and animals
Soil microbes play a major role in generating long-lived soil organic matter. Bacteria and fungi do this using enzymes, which generate polyaromatic compounds that are components of recalcitrant humic substances. The sink of carbon is under threat from climate change. This is because frequent wetting and drying cycles and elevated temperatures increase the accessibility of humic matter, raise enzyme activities, and stimulate microbial respiration resulting in more carbon dioxide evolution – and exacerbating the greenhouse gas effect. Among the numerous schemes for reducing atmospheric carbon dioxide is to persuade more soil microbes to carry out humification and generate recalcitrant carbon.
Where to now?
In a changing climate with increasing demands on land use, microbes are as much at risk as animal and plant communities. Soil microbial ecology is an essential component of macroecology and the below- and above-ground worlds are part of the same global environment. Does anyone know the rate of extinction of microbial genotypes in the tropics, the tundra, the rainforest or the Antarctic? Is microbial diversity disappearing at the same fast rate shown by plants and animals? Should we be storing and preserving, not only isolated microbial strains, but also soil samples? These would serve as reference points and help us evaluate the impacts of subsequent changes in soil use and management as well as climate change.
The 2015 International Year of Soils should remind the world that what we are standing on is not only growing our food, fuel and fibre but also maintaining the quality of our air and water. It is a precious resource that is under threat from human activities as well as commercial interests and political ignorance. Talking to other soil biologists is easy (we agree on the importance of our subject!) but it is making the connection between our science and national and international socio-economic policies and priorities that might win the day.
Half a millennium ago Leonardo da Vinci got it about right when he said, “We know more about the movement of celestial bodies than about the soil underfoot”: a thought echoed today by such luminaries as Julian Davies, Norman Pace and Craig Venter. Increasingly sophisticated molecular, microscopic, modelling and computational techniques are taking us closer towards untangling the complexity of the soil environment and revealing its many secrets. However, the ultimate practical goal is to manipulate our increasingly threatened soil resource to restore and maintain its crucial role in the biosphere and to enhance its capacity for food production.
RICHARD BURNS
School of Land and Food Sciences, The University of Queensland, Brisbane, Queensland 4072, Australia and Faculty of Science, Health, Education and Engineering, University of the Sunshine Coast, Maroochydore, Queensland 4558, Australia
[email protected]
[email protected]
FURTHER READING
A’Bear, A. D. & others (2014). Putting the ‘upstairs-downstairs’ into ecosystem service: What can aboveground-belowground ecology tell us? Biol Control 75, 97–107.
Bardgett, R. D. & van der Putten, W. H. (2014). Below-ground biodiversity and ecosystem functioning. Nature 515, 505–511.
Burns, R. G. & others (2013). Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol Biochem 58, 216–234.
Fierer, N. & others (2014). Seeing the forest for the genes: using metagenomics to infer the aggregated traits of microbial communities. Front Microbiol 5, doi:10.3389/fmicb.2014.00614.
Howe, A. C. & others (2014). Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci U S A 111, 4904–4909.
Krause, S. & others (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol 5, doi:10.3389/fmicb.2014.00251.
TerHorst, C. P. & others (2014). The relative importance of rapid evolution for plant-microbe interactions depends on ecological context. Proc R Soc B 281.
Image: Person standing in mud. Design Pics/Thinkstock. False-colour scanning electron micrograph of a root nodule on a pea plant. Dr Jeremy Burgess/Science Photo Library..
