Algal blooms and viruses
Issue: Water
17 November 2014 article

Most marine scientists have a love-hate relationship with dolphins, sharks, whales and turtles. They tend to get all the public's attention. We have pictures of them on our walls, own cuddly toys in their likeness and amuse our children with animated tales about their fictional adventures. Unfortunately, they distract from the real beauty and interest in the marine environment: the microbes, and in particular, the viruses.
Swimming with dolphins is a once in a lifetime opportunity for most people, yet whenever you take a dip in the sea you will immediately come into contact with billions of single-celled organisms and viruses. Since life evolved in the sea and crawled out onto land only a billion years ago, this microscopic community has an extra two billion years of evolution and genetic novelty over the metabolically dull dolphins and whales that crawled back in hardly any time ago.
The Earth is undoubtedly a blue planet, covered predominantly in water. We tend to think of the land plants as providing the oxygen we breathe, but in reality just as much oxygen comes from the single-celled, photosynthetic microalgae (known as phytoplankton) that float freely in our oceans. Yet most of the time our oceans don't look like pea soup, and that's because these algae get eaten by zooplankton, which then get eaten by fish, which then get eaten by bigger fish and so on.
However, the fate of phytoplankton is not limited to just being eaten: cellular destruction following viral infection also plays a major role.
Viruses are the most abundant biological entity on the planet, and can number in excess of 100 million in just one teaspoon of seawater. With an estimated 1031 viruses in the ocean responsible for 1021 infections every second, they are responsible for the destruction of up to half of the marine microbial community on a daily basis. This relentless lytic function is of fundamental importance to global biogeochemical cycling and ecosystem function, which would collapse without the constant recycling of nutrients at the microbial level.
Algae are highly opportunistic and, when conditions are right (for example, when a new source of nutrients becomes available, or when light and temperature become more favourable), phytoplankton species can swiftly come to dominate communities. These are known as algal blooms and are typified by single algal species reaching concentrations of over a billion cells per litre over large tracts of water bodies. Harmful red tides caused by toxic dinoflagellates such as Karenia brevis and Alexandrium fundyense are well-known examples of such community takeovers. In addition to causing water discoloration, they often produce toxins which can decimate fisheries and cause paralytic shellfish poisoning in humans. However, with all those viruses floating around and a relatively homogenous host population dominating in one region, viral infection becomes almost inevitable. Viruses have been found to be one of the major causes of bloom demise. Blooms offer a fantastic opportunity to isolate viruses. In July, a virus (AaV, 371 kbp, 377 genes) was described that infects the brown-tide-causing alga Aureococcus anophagefferens. AaV was estimated to be present at abundances as high as 1020 particles following a bloom.
Emiliania huxleyi
Perhaps the most famous of all the phytoplankton, the coccolithophore Emiliania huxleyi has global distribution and is well known for forming both coastal and open ocean blooms in temperate latitudes than can cover up to 100,000 km 2. As the most abundant and ubiquitous coccolithophore in modern oceans, it has become a key species for studies on global biogeochemical cycles and climate modelling. In a world obsessed with carbon footprints and emissions, E. huxleyi stands out as an organism capable of both fixing carbon dioxide, and of sequestering carbon in an inorganic form during the construction of its elegant chalk outer scales, known as coccoliths. Measuring just a few micrometres in diameter, but growing to high concentrations during times of blooming, over geological time, the release of its coccoliths to the ocean floor is capable of forming structures such as the White Cliffs of Dover. Amazingly, the death of these algae is even thought to influence the weather. When dimethylsulfoniopropionate (DMSP), which is used as an intracellular osmolyte, is released following cellular disintegration, it is rapidly cleaved to produce dimethylsulfide which rises up to the atmosphere, becomes oxidised and forms cloud condensation nuclei. Viruses will also help to accelerate all of these processes. Given the attention lavished upon E. huxleyi, it is unsurprising that coccolithoviruses have been isolated during E. huxleyi blooms. Indeed, the classical milky waters produced towards the end of E. huxleyi blooms (which can even be seen from space) are not full of healthy E. huxleyi cells at all; they comprise mostly dead and infected dying cells that have released their liths into the surrounding waters. When sampled, these milky waters are usually full of coccolithoviruses (EhVs).
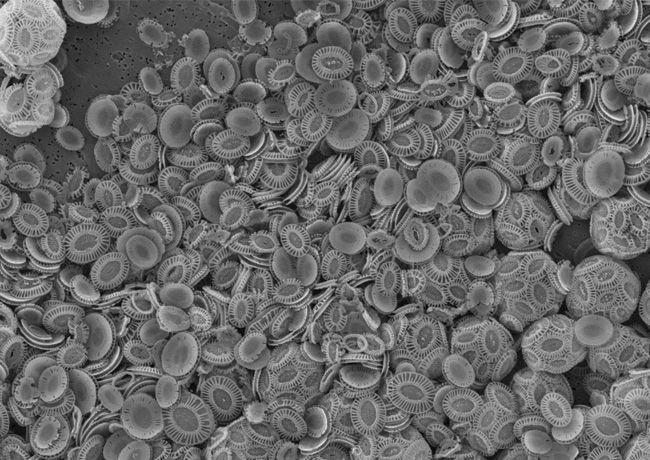
Giant viruses
Algal viruses have a profound impact on the global ecosystem. However, they have also gained attention for another reason: their size. The coccolithoviruses have been integral to changing the way we think of viruses. With gigantic genomes in excess of 407 kbp, these double stranded DNA viruses contain almost 500 genes. HIV manages to decimate the multicellular human body and its complex defence system with a mere nine genes in its genomic armoury. With nearly 500 genes and a single-celled host approximately five microns in diameter, the coccolithoviruses have revealed a complexity to viral infection that we had never observed before. Perhaps the most startling observation that can be made about EhV genomes is their genetic novelty: the vast majority of genes (>80%) are of unknown function and contain few database matches of note. The coccolithovirus genome, published shortly after its more famous cousin, the mimivirus (1,181 kbp, 911 genes), has opened our eyes to a new type of virus that seems to know no limits with regards to genomic size, content or complexity.
The term 'girus' (for giant-virus) was coined to refer to this new group of viruses, found within the nucleocytoplasmic large DNA virus family (NCLDV, a family which also includes the poxviruses). In the last decade, new viruses have been isolated including the Cafeteriavirus (730 kbp, 544 genes), Pithovirus (610 kbp, 467 genes), Sambavirus (1,213 kbp, 938 genes), Megavirus (1,259 kbp, 1,120 genes) and the recent Pandoravirus (~2,770 kbp, 2,556 genes). Each new giant virus has proven to be remarkably diverse; despite their shared evolutionary history, a mere dozen or so genes can be found in common between them. The Pandoravirus in particular is very interesting: despite its isolation in Acanthamoeba polyphaga, a host evolutionarily distant from the eukaryotic microalgae, its closest relative is the coccolithovirus.
There are certainly interesting times ahead in the study of algal viruses as their role in the decimation of algal blooms is understood with increasing clarity and the secrets of their genomes becomes unveiled. The major challenge marine virologists used to be concerned with was understanding and defining their impact in the global ecosystem; recent progress now suggests an understanding of their molecular modus operandi will prove a worthier challenge. Algal blooms are just the beginning of the story.
MICHAEL J. ALLEN
Plymouth Marine Laboratory, Prospect Place, The Hoe, Plymouth PL1 3DH, UK
[email protected]
FURTHER READING
Moniruzzaman, M. & others (2014). Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host-virus coevolution. Virology 466-467, 60-70.
Suttle, C.A. (2007). Marine viruses - major players in the global ecosystem. Nature Rev Microbiol 5, 801-812.
Wilson, W. H., Van Etten, J. L. & Allen, M. J. (2009). The Phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol 328, 1-42.
Yutin, N. & Koonin, E. V. (2013). Pandoraviruses are highly derived phycodnaviruses. DNA 516302773, 6.
Image: An example of a recent algal bloom in the Baltic. NERC Satellite Receiving Station, Dundee University. E. huxleyi (intact cells to the right) viewed under an electron microscope. Bar, 5µm. Mike Allen and Paul Rooks..
