A faster way to determine antimicrobial susceptibility
Posted on January 16, 2020 by Kerry Falconer
It can take as long as five days to determine which antibiotic treatments are likely to be effective in clearing a bacterial infection. These long wait-times can lead to ineffective antibiotics being used, delaying patient recovery and contributing to the increase of antimicrobial resistance. But is there a better way? In this blog, Kerry Falconer PhD student at the University of St Andrews discusses her research.
I am a third year PhD student currently working within the Infection Group at the University of St Andrews, supervised by Professor Stephen Gillespie. The group’s current research focuses on the detection and treatment of a wide array of infectious diseases from malaria to tuberculosis (TB). The aim is to improve the understanding, diagnosis and treatment of these challenging conditions through the development of new and rapid methods of detection and treatment.
As part of my undergraduate degree in Biomedical Science, I spent some time working in an NHS hospital. It was during this placement that I became interested in how research could help and improve patient care. My PhD has given me the opportunity to develop this interest further and to work in the field of clinical microbiology alongside infectious disease doctors, as well as clinical and research scientists within the NHS, industry and in academia.
One of the main problems faced in clinical microbiology is the inability to rapidly detect infections which are resistant to antimicrobials. This is especially problematic for time-critical conditions such as bloodstream infections, which are amongst the most serious and challenging investigations faced by clinical microbiology. Every hour treatment is delayed, the patient’s likelihood of survival is significantly reduced. Antibiotics continue to be the main choice for the treatment of bloodstream infections but the rise in antibiotic resistance has and continues to limit treatment options. It is therefore important to detect antimicrobial resistant infections quickly to ensure the patient is given optimal therapy at the first opportunity in order to resolve the infection effectively and to prevent further complications such as sepsis.
The need to administer antibiotic therapy quickly for bloodstream infections is hampered by the long turnaround times for bloodstream infection diagnosis and antibiotic susceptibility tests which can currently take 2–5 days. The main aim of my PhD is to investigate whether the current detection time of bloodstream infections and antimicrobial resistance can be shortened using a Scattered Light Integrated Collection (SLIC) device.
SLIC: A rapid antimicrobial susceptibility test device
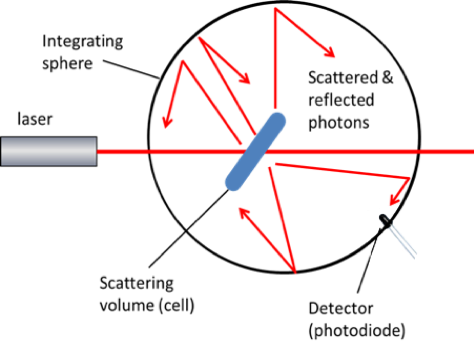
SLIC consists of laser, light collector and innovative electronics and allows the total light scatter to be analysed, enabling a very low bacterial concentration of 100 cells/mL to be detected.
Bacterial growth, inhibition and death can be sensitively monitored in real ,time using SLIC. When a bacterial population is treated successfully with an antibiotic, the number of bacteria cells will remain unchanged or will be reduced, and this can be detected on SLIC with a constant or reduced light scattering pattern. Alternatively, when the antibiotic is not effective, the bacteria will continue to replicate, the number of bacteria will increase and the light scatter detected on SLIC will increase. Therefore, SLIC is a valuable tool for differentiating between sensitive and resistant bacterial populations quickly and has so far been investigated for a number of clinical applications.
At the Federation of Infection Societies (FIS) 2019 Conference in Edinburgh I presented a poster on a recent clinical trial we conducted on the use of light scatter for the detection of bacteria in the bloodstream. The trial compared the accuracy and time of the present gold standard approach with SLIC for 101 patient samples. The study demonstrated the use of light scatter to be highly comparable to the existing gold standard approach and importantly allowed the time to detect antibiotic resistance to be shortened by more than 50%. This meant that it was possible to know what was causing the patient’s infection and importantly the most effective way to treat it on the same day. Notably, around 60% of samples processed contained antibiotic resistant bacteria.
The right antibiotic at the right time is crucial in ensuring a patient recovers from the infection. The early detection of infection and identification of the correct antibiotic to prescribe will greatly assist in patient care. Not only will it reduce the risk of treatment failures it will also promote good antibiotic stewardship, helping to ensure our existing antibiotics continue to be effective.

