A "superbug" with abnormal secretion system
Hello! I’m Sylvia Ighem Chi, a postdoctoral fellow in Dr Sandra Ramirez-Arcos’ group at Canadian Blood Services in Ottawa, Ontario, Canada. Dr Ramirez-Arcos and I would like to share with you some exciting findings of our recent research on the genomic characterization of the superbug Staphylococcus aureus.
S. aureus is naturally found in healthy humans, colonizing the respiratory tracts, skin and nasal passages. As an opportunistic pathogen, this bacterium causes diverse infections in communities and hospitals, including life-threating bacteraemia. In transfusion medicine, which is the focus of our research, S. aureus inhabiting the skin of blood donors can occasionally contaminate life-saving blood donations and derived products like platelet concentrates (PCs). This can happen during venipuncture or through breaches in sterility, which may lead to serious transfusion-transmitted reactions, including fatalities, in susceptible patients infused with S. aureus contaminated PC units.
The clinical symptoms manifested by patients after receiving PCs contaminated with bacteria depend on the patient’s immune status and the bacterial virulence factors. S. aureus is considered a major threat to patients due to its genome-encoded virulence determinants such as the type VII secretion system (T7SS), which plays a role in pathogenicity, survival and persistence in immune challenging environments. Six genes (esxA, esaA, essA, esaB, essB and essC) conserved across S. aureus strains form the T7SS core and are essential for protein transport.
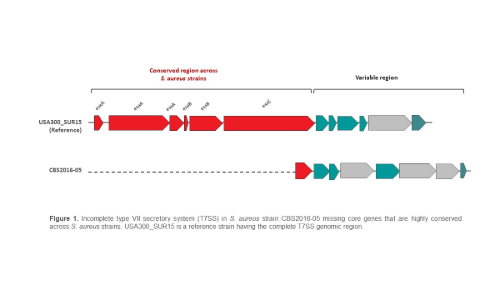
In our recent study, we performed a detailed analysis of the genomes of PC S. aureus isolates in comparison to other S. aureus genomes from blood and non-blood sources, with focus on virulence and antimicrobial resistance. Among the interesting findings, we found evidence of genome rearrangements involving a common genetic element Sa3int in two S. aureus strains from PC origin. This observation is not uncommon in S. aureus genomes. However, the most important finding reported in our study was the discovery of a truncated T7SS in a highly virulent S. aureus strain involved in a septic transfusion reaction of a Canadian patient in 2016 (S. aureus CBS2016-05). It was surprising to find that the CBS2016-05 isolate lacks this important virulence support system which is highly conserved across S. aureus strains (Figure 1). This strain probably compensates T7SS function by recruiting other export mechanisms and/or alternative virulence factors, such as neutralizing immunity proteins.
Our study reveals genome rearrangements in S. aureus isolated from PCs and reports the first S. aureus isolate lacking conserved T7SS core genes. These findings indicate that virulent S. aureus strains may have other mechanisms to fulfil the functions of the T7SS, which merits further investigation.
Image: Staphyloccocus aureus, MRSA - antibiotic resistant bacteria iStock/Artur Plawgo
