Behind the scenes of AppIndels.com
Can you introduce yourself, your role and your research area?
My name is Radhey S. Gupta. I am a professor in the Department of Biochemistry and Biomedical Sciences at McMaster University, Canada. My earlier research focused on the studies of mutants of mammalian cells, heat shock proteins (Hsp60 and Hsp70), and mitochondrial proteins. My interest in the evolutionary area was sparked by the serendipitous discovery of several prominent inserts/deletions (indels) in conserved regions of both Hsp60 and Hsp70 proteins. These conserved signature indels (CSIs: inserts or deletions flanked by conserved regions) had important implications regarding the evolutionary relationships between Bacteria and Archaea, origin of the eukaryotic cell(s), and the relationships between Gram-positive and Gram-negative bacteria. These studies, coinciding with the beginning of genomic era, showed me the usefulness of the rare genetic changes such as the CSIs for understanding/clarifying evolutionary relationships. Evolutionary relationships and classification of microorganisms in late 90s, and even now, is primarily based on branching of species in the 16S rRNA, or other genes/proteins trees. For most prokaryotic taxa, no characteristic is known that is uniquely shared by all species from a given group/taxon.
In a 2002 review, “Critical Issues in Bacterial Phylogeny”, we surmised that for a more reliable/informative understanding of microorganisms it is essential that well-defined means should be identified for the demarcation of different taxa. Taxon-specific CSIs provide trustworthy means (i.e. synapomorphies) for this purpose. Hence, for the past >25 years, a major focus of my research group has been on identifying CSIs specific for different groups of organisms. These studies have identified >2000 CSIs specific for >250 prokaryotic taxa, and they are proving instrumental in clarifying the evolutionary relationships and classification schemes for several important groups of microorganisms.
Tell us about your most recent paper?
Our current publication describes the development of a web-based tool/server (AppIndels.com) that uses the sequence information for previously identified taxon-specific CSIs for predicting the taxonomic affiliation of a genome. The server has a very simple user interface requiring no expertise to use or interpret the results. For any submitted genome, the server performs analysis. If the submitted genome contains significant number of CSIs specific matching a particular taxon, then the name of that taxon along with the total numbers of matching CSIs, along with sequence information are displayed.
The working of this server is based on the observation that the CSIs for different taxa, identified in earlier work, exhibit a high degree of predictive ability to be found in other members of these taxa. Based on this property, information regarding the presence of known taxon-specific CSIs in a genome can be utilized for predicting its affiliation to a specific taxon. However, there was no convenient method/tool available to perform such analysis. The AppIndels.com server (and its predecessor - Indels.com) was developed with the aim of filling this gap, by creating a simple to use method/tool, which can readily interrogate the presence of known CSIs in a genome sequence for taxonomic/diagnostic purposes. A proof-of-concept of using sequence information for known CSIs for predicting taxonomic affiliation was obtained by the creation of Indels.com server in 2018 with the assistance of Joseph Manalo (a computer science undergraduate student). However, limited work was carried out on its validation. To undertake further work on its validation, David Kanter-Eivin (an undergraduate student in the Arts & Science program) was hired in May 2021. David made several modifications to the Indels.com server to make it more robust in performance and introduced a weight criterion to account for the differences in the number of CSIs known for different taxa. The updated server was hosted under the name AppIndels.com.
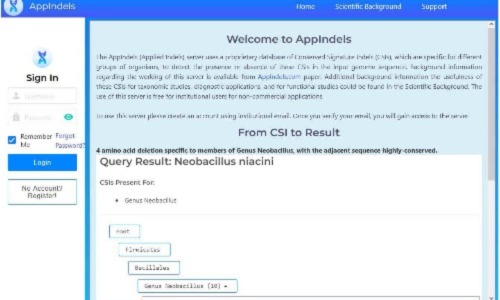
The present paper describes detailed studies on the working and validation of the Appindels.com server. For validating the performance of this sever, sequence information for Bacillus spp. was used. Species from the genus Bacillus were recently reclassified into >30 different genera, each of which could be reliably demarcated based upon multiple identified CSIs. Besides the named Bacillus species, which were reclassified into different genera, genome sequences were also available for >720 Bacillus spp. whose taxonomic affiliation was unclear. For testing the performance of AppIndels.com server, we used a database of 585 CSIs, which included >350 CSIs specific for ≈ 45 Bacillales genera. The server’s performance was examined by testing its predictions for the 721 Bacillus spp. of unknown taxonomic affiliation. Based upon the presence of CSIs specific for different genera, the server predicted that 651 of these Bacillus spp. corresponded to 30 Bacillales genera. Phylogenetic analyses confirmed that all predictions made by the server were correct. The results obtained demonstrate that the AppIndels.com provides a useful new tool for the identification of known taxon-specific CSIs in genome sequences and using this information for taxonomic and other applications.
What is your research about and why is it important?
The AppIndels server is unique from all other methods (using phylogenetic analysis or genomic relatedness) for inferring taxonomic affiliation or evolutionary relationships. Unlike other methods, the taxon affiliation by this server is based upon the shared presence of multiple CSIs which are specific for a particular taxon. The results from the server thus should aid in the demarcation of different prokaryotic taxa, based on uniquely shared molecular characteristics. Based on earlier work, the CSIs in genes/proteins sequence have been shown to play important (or essential) functions in the organisms for which they are specific. As the genotype specifies phenotype, biochemical studies on understanding the cellular functions of the identified CSIs, which are specific for different taxa, should lead to discovery of novel characteristics, which are specific for different groups of organisms. Additionally, the sequences of these taxon-specific CSIs, which are present in conserved regions, also provide important means for the development of novel diagnostics, and potential therapeutics.
One limitation of the AppIndels.com is that it can predict taxonomic affiliation for only those taxa for which CSIs have been identified and present in its database. To enhance its utility, it is important to enlarge the CSIs database. The CSIs specific for several other prokaryotic taxa have been previously identified and information for them will be added to the server’s database upon completion of validation studies. Additionally, our future work will focus on identifying CSIs specific for other important groups of prokaryotic organisms.
You can read Professor Gupta's full article in 'AppIndels.com server: a web-based tool for the identification of known taxon-specific conserved signature indels in genome sequences. Validation of its usefulness by predicting the taxonomic affiliation of >700 unclassified strains of Bacillus species' in International Journal of Systematic and Evolutionary Microbiology.
