Delving into the Diversity of Fusobacterium nucleatum
Hey there! My name is Claire Crowley, I am a final year PhD student in Microbiology at the Dublin Dental University Hospital, Trinity College Dublin, Ireland. As a member of the oral microbiome research group, headed by Dr. Gary Moran, I undertook my research on the oral bacterium Fusobacterium nucleatum.
The oral cavity is one of the most diverse habitats in the human body, with ~700 species of bacteria, plus yeasts, viruses and some protozoa which are collectively referred to as the oral microbiome. This collection of microbes can shift and change over the life-course and can be influenced by many factors including age, diet, tobacco consumption and disease. Some of the most common microbiome-related diseases in humans are caused by changes to this microbiome, including dental decay (caries) and gum diseases (gingivitis and deeper periodontal disease). Your daily routine of brushing and flossing can be considered “microbiome-management” and helps prevent these common oral conditions.
An important member of the oral microbiome is F. nucleatum. This bacterium plays a key role in the formation of the plaque biofilm on tooth surfaces. It acts as a bridging organism between species which are early colonisers of the tooth surface, such as streptococci, and late colonisers, including Porphyromonas gingivalis. F. nucleatum, in combination with P. gingivalis, is often associated with gingival inflammation and gum disease in the oral cavity. More recently, however, research has begun focusing on the potential for F. nucleatum to act as an oncobacterium, specifically in the context of colorectal cancer, where increased abundance of F. nucleatum can be found on tumour sites compared to healthy colorectal tissue. Furthermore, the presence of F. nucleatum at these tumour sites has been associated with poorer patient outcome and chemotherapy resistance.
Previous work carried out in our lab focused on the study of bacterial species present on oral leukoplakias (OLK). These are abnormal white patches found inside the mouth which have the potential to transform into oral cancer. From our research, it was found that a greater relative abundance of F. nucleatum was detected on these OLK sites, compared to healthy contralateral sites in the same patient. This information, coupled with the association between F. nucleatum and cancer, led to deeper investigations into genomes of the F. nucleatum isolates which are found on these potentially malignant sites, to discern whether they may play a role in the progression of oral disease.
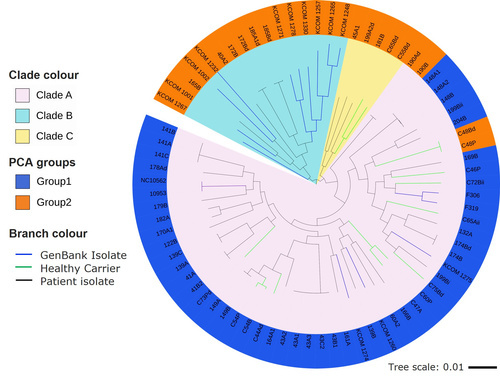
Firstly, isolates were recovered from OLK sites and healthy sites of patients attending clinics at the Dublin Dental Hospital. These swabs were plated on selective media to aid in the isolation of Fusobacterial species, which were confirmed by sequencing of the 16S rRNA gene. Using this method, we found that the most commonly isolated Fusobacterium was F. nucleatum subsp. polymorphum. To analyse the genetic relatedness of these isolates, we decided to carry out whole genome sequencing using short-read Illumina sequencing. From this we carried out pangenome analysis to determine the core genome (genes present in all isolates) and accessory genome (genes that are present in less than 95% of the isolates). This analysis revealed a small core genome of 1,604 genes. Based on the alignment of these shared, core genes, a phylogenetic tree was generated that divided the isolates into three genetic clades. However, we found no association with disease severity and phylogeny. It was interesting to note that isolates from the same patient, whether from a healthy site or an OLK site, often clustered together on the tree, indicating that these were genetically closely related, and suggested there was no malignancy-associated genotype.
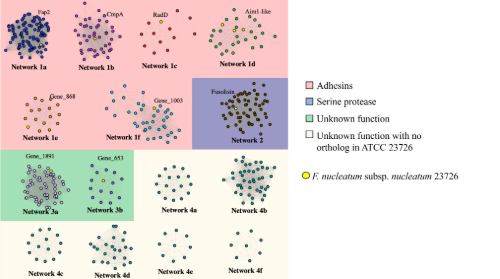
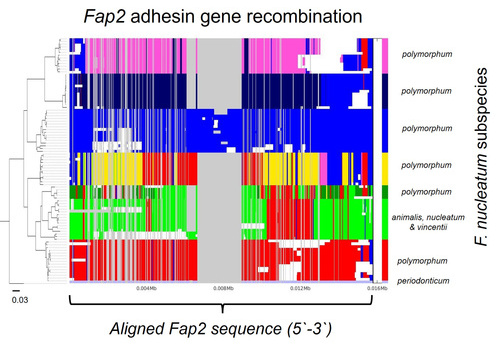
Outside this shared, core genome, we found a much larger accessory genome of 6,032 genes, suggesting that the larger part of the genome was fluid and probably shaped by horizontal gene transfer. Interestingly, within this accessory genome we discovered a large number of genes encoding Type Va adhesins, a family of outer membrane adhesins implicated in virulence and F. nucleatum-host-cell interactions. These genes showed great heterogeneity in their distribution across the isolates, with some isolates possessing only one of these adhesins, and others possessing as many as eight. Adhesion assays carried out on oral keratinocytes, with isolates that possessed different numbers of adhesins showed varied ability to adhere to these cells, with isolates possessing fewer adhesins tending to display weaker adhesion phenotypes. Most surprisingly, alignment of these adhesin gene sequences from different F. nucleatum subspecies, namely animalis, nucleatum, polymorphum and vincentii showed that they exhibited genetic mosaicism as a result of high levels of recombination. This included the well characterised cellular adhesin Fap2 and the aggregation factor RadD.
This work highlights the genetic heterogeneity present in isolates of F. nucleatum, especially in the adhesins it possesses, and opens up many exciting avenues of investigation. It is important, in future, to continue to characterise these isolates and the genes they possess to fully understand F. nucleatum pathogenesis and elucidate the role it plays in the progression of disease. Our ongoing analysis is showing that this genetic heterogeneity is reflected in how different isolates interact with malignant oral cells, suggesting that some genotypes may be more oncogenic than others. A deeper, more complete view of F. nucleatum may aid in the development of therapeutics against the bacteria, to improve the outcome of patients with conditions from periodontitis to colorectal cancer.
Image: iStock/Inventori
