From autoimmune disease to taxonomy: a positive outcome from COVID-19 Lockdown
Posted on November 23, 2023 by Professor Emerita Sheila Patrick and Dr Linda Stewart
Professor Emerita Sheila Patrick and Dr Linda Stewart take us behind the scenes of their latest publication 'Genomic analyses of Bacteroides fragilis: subdivisions I and II represent distinct species' published in Journal of Medical Microbiology.
In 2019 Jamie English embarked on his laboratory-based PhD project under our supervision. The plan was for him to study a protein produced by a bacterium, Bacteroides fragilis, which resides normally in the human gut. This protein, called BfUbb, is highly unusual. It mimics the shape of a protein called ubiquitin which is found in all human cells, but not in bacteria.
The gene for the protein had been discovered during the first complete genome sequencing project of B. fragilis in the early 2000s. Professor Sheila Patrick was the Principal Investigator on the genome project and along with Professor Garry Blakely from the University of Edinburgh, Scotland, studied BfUbb. It is an intriguing protein; the amino acid sequence and shape of the protein are similar to the human equivalent; however, unlike the human equivalent it doesn’t function inside the cell but is exported outside the cell associated with outer membrane vesicles.
Dr Linda Stewart had studied B. fragilis as a PhD student of Professor Patrick in the late 1980s in quite a different context. B. fragilis lives quite happily contained in the gut, but if the gut wall is breached, for example if appendicitis causes a rupture, faeces escape into the abdomen and B. fragilis can cause life-threatening infection. Antibiotics are administered to people having gut surgery to prevent this type of infection.
After starting her career as a Biomedical Scientist in a clinical microbiology laboratory, Dr Stewart returned to Queen’s University Belfast, Northern Ireland, becoming a lecturer in 2013 and had gained considerable recognition for her expertise in phage technology and immune detection in relation to tuberculosis. Professor Patrick asked if she would be interested in collaborating on the BfUbb project to test the hypothesis that, as BfUbb is so close to human ubiquitin in shape, it could be a trigger of autoimmune disease.
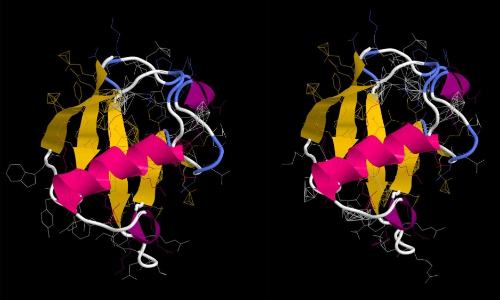
The starting point was to analyse serum from people being tested for a number of immunological conditions. We were able to detect antibodies to BfUbb in human blood serum samples. So, although the bacterium lives in the human gut, BfUbb can generate an antibody response. People with autoimmune disease are more likely to have these antibodies than healthy volunteers and some people have antibodies that bind to both BfUbb and human ubiquitin.
The plan for Jamie’s PhD was to investigate this potential link between autoimmune disease and antibodies to BfUbb in Dr Stewart’s Laboratory.
Then COVID-19 came along, and laboratory work was not possible due to lockdown.
We approached Professor Lesley Hoyles from Nottingham Trent University, UK, an expert in bioinformatics and an experienced taxonomist, to see if she would be willing to collaborate and teach Jamie how to go about bioinformatic computational analyses, initially to look for the BfUbb gene in publicly available B. fragilis genomes.
With the help of Professor Hoyles and Dr Newbury, a post-doc in Nottingham Trent University, UK, the research diversified to include phylogenetic analyses, characterisation of the antimicrobial resistance (AMR) genes and analysis of the pangenome. Since the 1970s B. fragilis has been divided into two groups, I and II, based on DNA-DNA homology. Our comparison of both the core and accessory genes in the genomes, now published in Journal of Medical Microbiology indicates that Division II B. fragilis (referred to B. fragilis A in genome databases) can be considered to be a separate species, distinct from Division I B. fragilis.
Screening of antibiotic genes in the genome sequences (the resistome) confirmed that all B. fragilis Division I genomes encoded cepA, an AMR gene that was absent in the Division II B. fragilis A genomes analysed. This gene confers resistance to some beta-lactam (penicillin and related) antibiotics by production of a beta-lactamase enzyme that breaks them down. In contrast, all B. fragilis A genomes contain a gene for another type of beta-lactamase enzyme, cfiA/ccrA, that breaks down different types of penicillin-related antibiotics, in particular carbapenem.
Our science indicates that Division II Bacteroides fragilis should be a new species, with a different name. But changing the names of bacteria based on scientific evidence can have unexpected consequences for practical bacteriology. This is particularly important in relation to bacteria that cause infection; an unfamiliar name can result in a wrong diagnosis and inappropriate treatment.
Bacterial nomenclature change proposals should consider the potential impact of name change. Will it be helpful to, for example, the clinical microbiologist? Will the name change be easily memorable and relate to previous names to allow understanding of the infectious context?
So, although the science points to a new Bacteroides species, is a name change appropriate and helpful?
We think so and have proposed that Division II Bacteroides fragilis A is renamed Bacteroides fragila. This will highlight the different antimicrobial susceptibilities and ensure that the clinical association with the potential for lethal infection arising from these bacteria remains easily memorable. We are currently working to compile the taxonomic details required for a formal proposal with approval from clinical colleagues.
And what of BfUbb?
There are still many unanswered questions, not least what are the potential health implications of having antibodies that cross-react with human ubiquitin circulating in the body? Inflammatory bowel disorders, autoimmune disease, stroke and fertility are just some examples of where this might be important. Ubiquitin is involved in the function of all the cells in the body, so there are potentially many more. The biggest issue for any area of scientific endeavour is to convince reviewers that research of an unusual and unexpected finding is worth funding, exemplified by Dr Katalin Kariko who was awarded the 2023 Nobel Prize for Medicine. We have had multiple rejections over the years. We haven’t yet given up!

