Link between antibiotic resistance and prophages
My name is Helena Leinweber and I performed my PhD studies as a Marie Curie fellow in Professor Hanne Ingmer’s research group at the Department of Veterinary and Animal Sciences of the University of Copenhagen, Denmark. Here we study food borne pathogenic bacteria and zoonoses that are bacteria transmitted from animals to humans. We aim to provide new knowledge relevant to prevention, control and treatment of antibiotic resistant bacteria including alternatives to antibiotics, phage therapy and hygiene.
Antibiotic resistance is a global public health crisis, and gaining insights into the relationship between resistance mechanisms and other genetic elements, such as phages, is crucial for effective antibiotic resistance management. This study came about when our collaborators at Statens Serum Institut, the Danish Center for Disease Control, noted a curious correlation between fluoroquinolone-resistant Staphylococcus aureus isolates and phage carriage. So we decided to investigate further.
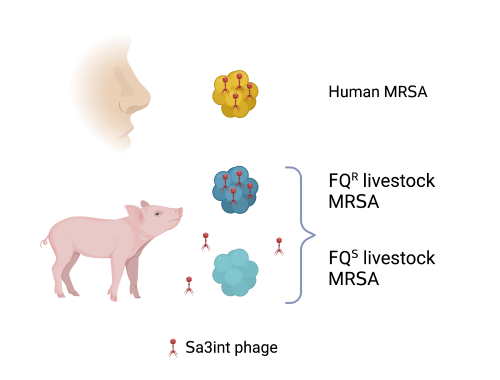
Research on S. aureus is particularly important as it colonizes about a third of the human population in addition to a wide range of animals and has the potential to cause a wide range of diseases. The preference of staphylococcal strains for human or animal hosts is in part dependent on content of mobile genetic elements. A prominent example of the latter are prophages of the ΦSa3int family, which are bacterial viruses that can stably integrate into the DNA of S. aureus, which is then called a lysogen. Those specific phages are found in most human strains of S. aureus where they express one or more so called immune evasion factors, shown to facilitate human colonization as well as to promote human-to-human transmission. In contrast, livestock-associated methicillin-resistant S. aureus (LA-MRSA) commonly lack those phages. However, there have been reports of LA-MRSA isolated from humans and such transfer events are often associated with Sa3int prophage carriage. This could potentially lead to difficult to treat infections and gaining knowledge about the mechanism behind these transfer events could help to control the spread of antibiotic-resistant pathogens and protect human health.
Back when our colleagues collected the LA-MRSA strains harboring ΦSa3int prophages they noticed that all of them also were resistant towards fluoroquinolones (FQ). This caught our attention as the use of FQ is tightly regulated in Danish food production animals. We thus wondered whether there may be a link between prophage carriage and FQ-resistance.
For our study we introduced FQ-resistance mutations into a laboratory strain of S. aureus that mimics a LA-strain. We compared the behavior of the FQ-resistant mutant strain with the FQ-susceptible wild-type strain in terms of phage integration and release. Phage Φ13, a representative of the ΦSa3int phage family, was used for monitoring.
In a first round of so called lysogenization assays which monitor the rate of phage integration into the bacterial chromosome, we did not find any significant difference between the FQ-resistant mutant strain and the wild-type strain. In a second experimental setup we then examined whether the FQ resistance mutations influenced spontaneous or mitomycin-induced release of phage Φ13 from lysogens (bacteria carrying the integrated prophage) but found no significant difference in phage release either. The study's findings indicate that the mutations associated with FQ-resistance in LA-MRSA strains do not contribute to the presence of ΦSa3int phages. This suggests that other factors may be involved in the acquisition and integration of these phages in LA-MRSA strains.
We do however acknowledge that our study has some limitations. One limitation is that the model phage and bacterial host applied when assessing phage-bacterium interactions differ from the bacteria and actual phages present in our collected strain. We suggest that future experiments should address the interactions between the collected phages and their natural S. aureus hosts. Understanding the factors contributing to the presence of ΦSa3int phages in LA-MRSA strains can help assess the likelihood of transmission and identify potential strategies for prevention and control.
I myself have moved on from Hanne Ingmers lab after my PhD but I really hope a future student will keep working on this project as this could provide important information for understanding this process. Also, I am super happy to see this study published now, especially as it has been quite challenging to continue the work on the manuscript a year after I defended my PhD…
We would like to thank Microbiology Society for also giving space for “negative results” and thereby promoting transparency, avoiding publication bias, and fostering a culture of openness!
Image: Md Saiful Islam Khan
