Nanopods: Hardwiring in bacterial biofilms
My name is William Hickey, I’m Professor Emeritus at the University of Wisconsin-Madison in the Dept of Soil Science. Research in my lab centres on elucidating metabolic mechanisms that underlay bacterial biodegradation of environmental chemicals. Our recent paper in Access Microbiology, which focuses on the plant pathogen Paracidovorax citrulli, is an outcome of the serendipitous discovery of nanopods, made as part of a biodegradation project. In 2008, we were using transmission electron microscopy (TEM) to examine cells of the bacterium (Delftia acidovorans) for changes in envelope structure after biodegradation, and observed unusual structures in the culture fluids (Fig. 1). The identity and nature of these intriguing structures was unknown to us, as well as all of the microbiologist colleagues could identify. Thus, began the quest to determine the structure of these nanopods, as the in TEM image gave the appearance resembling pea pods. These intriguing structures were something that neither we, nor any of our microbiologist colleagues, could identify. Thus, we began the quest to unravel the mystery of nanopods.
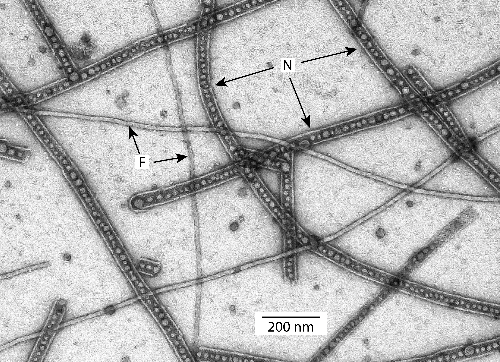
Nanopods in culture fluids of Paracidovorax citrulli imaged by negative staining and transmission electron microscopy. Nanopods are indicated by “N”, flagella are indicated by “F”. Spherical structures within nanopods are outer membrane vesicles.
We employed a battery of biochemical, genetic and physical methods to elaborate the structure and function of nanopods. Collectively, these experiments revealed that nanopods consisted of outer membrane vesicles encompassed by a surface layer protein (SLP) that we named Nanopod Protein A (NpdA). Studies with D. acidovorans showed nanopod formation was inducible and mutants deficient in nanopod production leaked metabolites, suggesting a physiological connection. Most surprising were results obtained by imaging of thick (ca. 1250 nm) sections of intact biofilms by three-dimensional scanning electron microscopy (3D-SEM). These images revealed nanopods existed natively as block-like extensions protruding from cell surfaces (Fig. 2). Frequently, nanopods were contiguous with surfaces of two adjacent cells with portals opening from the base of the nanopod into the cells. Thus, nanopods presented the appearance of structures potentially capable of mediating intercellular transmission of materials. Nanopod networks were unprecedented structures and presented the possibility for materials to be directly shared between cells.
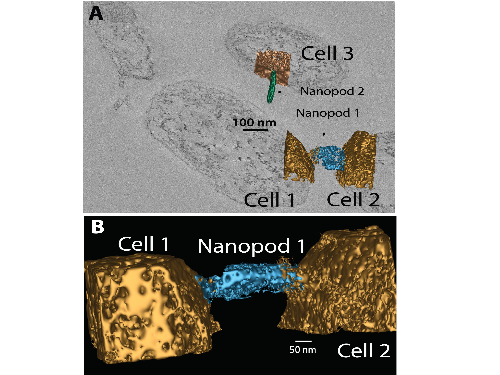
Our report in Access Microbiology built on this body of knowledge and extends research on NpdA and nanopods beyond the field of biodegradation. One new research area is plant pathology, and our report established that the surface layer (S-layer) formed by NpdA is a key characteristic of Paracidovorax citrulli (causative agent of Cucurbit Blotch Disease) and a number of other plant pathogenic bacteria. For bacterial pathogens of animals, many different types of S-layers are known and these often have key roles in disease development. Thus, our study in Access Microbiology provides the first report of an S-layer in plant pathogens and opens the door to future research examining the role of S-layers in virulence of these organisms. Another new area opened for NpdA and nanopod research is marine microbiology, as our study showed that a number of marine bacteria possess an NpdA-based S-layer. Interestingly, one of these marine bacteria is Nitrosococcus oceani, which was one of the organisms in which S-layers were first visualized by freeze-etching in the 1960’s. However, the SLP composing the N. oceani S-layer has never been identified; based on our data in our Access Microbiology, an NpdA ortholog is a strong candidate.
Perhaps most the intriguing finding in our Access Microbiology report is the observation of nanopod networks in P. citrulli biofilms similar to those documented for D. acidovorans. Thus, our work so far indicates that formation of nanopod networks is a regular feature of bacteria forming an NpdA-based S-layer. The key goal now is to determine whether or not intercellular transfer of material does occur through nanopods. If so, the impacts could be widespread and necessitate re-evaluation of some concepts in microbial physiology and ecology. More broadly, it’s possible that formation of nanopod networks may not be limited to bacteria with NpdA-based S-layers, and may occur in bacteria with S-layers composed of other SLPs. For example, nanopod-like fragments have been documented in cultures of Caulobacter crescentus, which produces an S-Layer composed of RsaA. Thus, examining biofilms of C. crescentus by TEM (ideally by 3D-STEM) for evidence of nanopods would be another interesting avenue of future work.
Of all the work I’ve done in microbiology research over the past 30+ years, the nanopod project has been the most interesting and rewarding. I encourage others to follow their instincts and pursue their own serendipitous findings especially when these diverge from the well-travelled paths of popular research topics.
Image: William Hickey
