One C. psittaci clone to rule them all?
Posted on October 24, 2022 by Martina Jelocnik
Upon the publication of her team's latest paper in Microbial Genomics, Dr Martina Jelocnik gives a behind-the-scenes look at the work of the University of the Sunshine Coast's Chlamydians team, keeping track of veterinary chlamydial infections.
Tell us a bit about yourself and your work.
I am Dr Martina Jelocnik, a veterinary microbiologist from the Centre for Bioinnovation at the University of the Sunshine Coast (UniSc), Australia. As a Research fellow, I lead the UniSC molecular “Chlamydians” team, investigating key questions in molecular epidemiology and developing diagnostics for 'One Health' and other veterinary chlamydia. I am also a very enthusiastic microbiology educator.

UniSC Chlamydians and kangaroos on the UniSC Sippy Downs campus. From the left: Vasilli Kasimov, Susan Anstey, Rhys White, and Dr Martina Jelocnik.
What is your research about and why is it important?
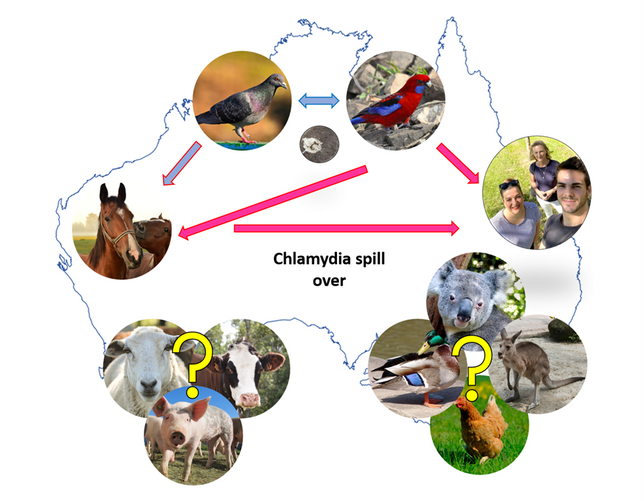
In Australia, the bacteria Chlamydia is best known as the notorious pathogen that causes devastating disease in koalas. But besides koalas, many other native (such as parrots) and domesticated (like horses and livestock) animals also become infected with Chlamydia. In particular, Chlamydia psittaci (C. ps), poses a threat to human health, but also to horses. With other “Chlamydians” across Australia, we described the unusual cases of C. ps in horses, resulting in reproductive loss and zoonotic transmission to humans. While the C. ps infections in native parrots (and Australians in the case of zoonotic infections) are well known, the widespread descriptions of C. ps in thoroughbred and other horses were considered novel and raised biosecurity concerns. Chlamydial horse infections occur every year across Australia. Also, these horse infections are currently considered Australian phenomena, as there are no (or scarce) reports of horse C. ps infections elsewhere.
Using limited whole-genome sequencing (WGS) and molecular typing, our “One Health” approach, intertwining industry, wildlife and horse veterinarians, diagnostics laboratory and other researchers, revealed that human, parrot and horse C. ps strains are genotypically identical. So, we wanted to expand the available horse C. ps genomes, try to figure out ’who is giving it to who’ and when these modern strains may have appeared in Australia.
What does a normal day in the lab look like?
We are like Chlamydia detectives, tracking its movement to understand the ‘where, when and how’ of infections and identifying the usual and unusual ‘suspect strains’. For example, we ourselves and/or broader Australian and global Chlamydians (co-authors on our most recent study) detect C. ps in horses, birds or other hosts; or identify a ‘C. ps suspect’; and then pass it to us for phylogenetic and sequence analyses. But here is the catch, in Australia C. ps is classed as a Physical Containment level 3 (PC3) strict intracellular organism, making it hard to grow in the regular research and/or diagnostic PC2 lab. The PC3 facility must meet the specified design and construction requirements. As intracellular pathogens, chlamydia only grows inside eukaryotic cells, additionally requiring cell culturing. Furthermore, due to delays in sample transport or inappropriate sample storage, it is sometimes hard to isolate live and viable chlamydia from clinical samples. So, we used culture independent probe-based WGS to ‘fish out’ C. ps in samples, in addition to molecular typing.
What are your findings so far?
We found that parrot and horse strains are clonal, meaning they are both descended from a single progenitor strain, but we could see fine differences between the C. ps clones. We also observed that we could have diverse strains, such as those found in Australian native pigeons. Finally, we think that the parrots are spilling C. ps over to horses and that these modern clonal strains have existed in Australia at least since the 1960s.

What do you enjoy most about your research?
I love working on C. ps (and other novel and traditional Chlamydiae) because they remain so enigmatic. In Australia, we report limited genetic diversity and host range compared to that globally. Our colleagues, the “global Chlamydians”, provide exciting and significant reports of genetically diverse C. ps infecting such a wide range of wildlife and domesticated animals, and, most importantly, humans. We always wonder whether we are missing some hosts/reservoirs. In Australia (and elsewhere), these infections cause economically significant diseases and pose risks to human health. We continue to track C. ps, as it remains a bit of a mystery who else has it (besides horses, parrots and humans) and what strain it is. This is very important (and interesting) in understanding the epidemiology of the enigmatic C. ps.
What do you hope the future implications of your research will be?
I hope that this and other chlamydial research continues to be supported as it provides a nexus between researchers, government and industry to effectively manage One Health chlamydial infections, and importantly, reduce the risks to humans. In addition, this will allow for the development and wider use of rapid Chlamydia diagnostics, and vaccines to prevent disease.
