Prophages: An understated part of our microbiome
My name is Laura Inglis, I’m a PhD student in Robert Edward’s lab at Flinders University, Australia. Our lab does a lot of metagenomic work, looking at clinical and environmental microbiomes. My research is particularly focused on the phage component of the human microbiome.
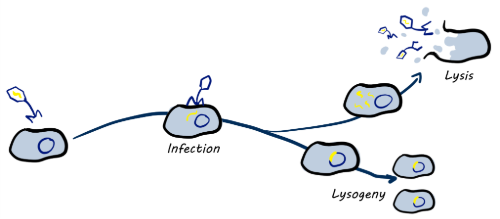
Phages are viruses that infect bacteria and are very host-specific, sometimes only infecting a handful of strains in a species. Phages have two main lifecycles; lytic phages which infect, replicate and destroy their hosts, and temperate phages which will instead integrate into their bacterial hosts DNA until changes in conditions trigger a switch to the lytic cycle.
While integrated into their hosts they are referred to as prophages and the combined host-prophage cell is called a lysogen. These prophages can provide the lysogen with benefits such as projection from lytic phages and toxin production genes. A few well-known pathogens only cause symptoms when they have their prophage that provides the toxin production genes. Shiga toxin in E. coli and cholera toxin in V. cholerae are two such examples.
Despite this, prophages are still a relatively understudied part of the human microbiome, so we decided to look through genomes uploaded to online databases to get as wide of an overview of prophage rates in the human microbiome as possible.
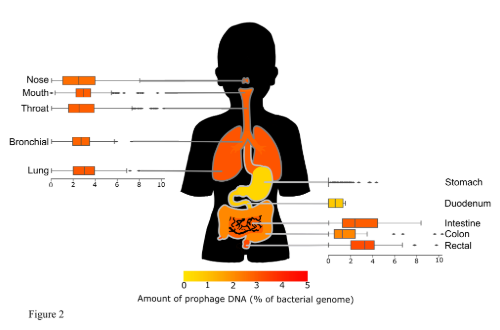
Prophages were thought to be pretty ubiquitous throughout all species of bacteria and we largely found this to be the case, with prophages making a sizeable percentage of most bacterial genomes. However, the specific amount of prophages varies significantly across various factors. Area of the body the bacteria were samples from was a major factor, with some areas like the stomach having almost no prophages, potentially due to the uniquely harsh environment. Another factor was isolation region, it is known that the gut microbiome is affected by location and we found that the amount of prophages followed suit. The health of the human host was also a factor that showed significant differences, but only once body site was taken into account. In some areas of the body the pathogenic bacteria sampled from symptomatic people had a higher amount of prophages than the bacteria samples from healthy individuals.