This toolbox could use a flashlight: adapting split-luciferase as a new tool for detecting proteins secreted by Bacillus subtilis
Microbiologists study lifeforms that usually cannot be seen by the naked eye. For molecular microbiologists the study objects become even smaller, like DNA, proteins and metabolites. How can we study what we cannot see?
My name is Mariah Kes. I am a PhD student at the Department of Molecular Microbiology at the Vrije Universiteit Amsterdam, The Netherlands. Detecting proteins is an important part of my research project. I study the bacterium Bacillus subtilis, which is used in the bioindustry to produce and secrete recombinant proteins into the culture medium from which they can be easily purified. B. subtilis is an attractive host for this purpose, because it is safe to use and known to secrete proteins across its single membrane efficiently.
My goal is to improve the production levels of recombinant secreted proteins. Therefore, I need to be able to detect and quantify the protein of interest in the culture medium.
Sure enough, there are already many tools available in the protein detection toolbox. However, we wished to take a new approach to create a fast, sensitive and quantitative protein detection method that is independent of the studied protein. In our recent paper, my colleagues and I set up a detection method based on luciferase. This is an enzyme that catalyzes a chemical reaction in which light is produced, like a tiny molecular flashlight. By attaching this luciferase flashlight to a protein, the luciferase can act as a reporter for the presence of the protein. If we detect light, we know the protein is there.
However, adding a complete flashlight to secreted proteins may cause problems. The secretion system translocates proteins in an unfolded state, and adding a possibly folded, bulky luciferase may clog the protein conducting channel in the membrane. Therefore, we chose to use a split-luciferase, which is a luciferase divided into two parts: a small peptide and the remaining bulk part of the protein. So, we took away the battery from the flashlight, resulting in two parts that cannot function on their own. The battery, representing the peptide, can be attached to the protein of interest and should not influence its secretion. When combined with the remaining flashlight, the larger part of luciferase, in the culture medium, they reassemble into a functioning flashlight.
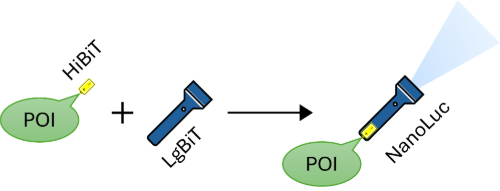
Schematic overview of the split-luciferase assay. When HiBiT, tagged to a protein of interest (POI), is mixed with LgBiT, they assemble into a functioning NanoLuc luciferase.
The split-luciferase we used in our project is called NanoBiT, which is based on the luciferase NanoLuc. This is a small and optimized luciferase that was developed by researchers from Promega Corporation and Promega Biosciences Incorporated. The two split-luciferase components are called HiBiT, the small peptide, and LgBiT, the remaining luciferase.
The project started as a research project for an undergraduate student. It was her task to figure out whether the split-luciferase assay would work for the detection of proteins secreted by B. subtilis. We assumed that at high production levels, we would have the highest chance of detecting it with the new assay. Therefore, we chose to start with a strongly expressed protein: the native xylanase A (XynA).
We quickly found out that we could indeed detect the secreted XynA-HiBiT proteins, but also learned that at these high production levels the luminescence signal was not quantitative. High HiBiT levels deplete the LgBiT and substrate quickly, resulting in an underestimation of the production levels. It felt ironic that we aim to develop a protein detection method to eventually improve production levels, but the levels were already too high for the new method to function! Soon, we realized that we needed to dilute our protein samples to obtain luminescence values that truly reflect the produced protein levels.
After the student’s successful effort to adapt the split-luciferase system for the detection of proteins secreted by B. subtilis, we continued developing a medium throughput assay to test multiple strains and conditions simultaneously. We were successful in monitoring the secretion over time of three other HiBiT-tagged heterologous proteins: amylase M (AmyM), protein glutaminase A and a nanobody targeting GFP. We were successful in monitoring the secretion of all proteins over time. Furthermore, we tested the assay to find improved protein secretion conditions. Specifically, we changed the signal peptide of AmyM and used the split-luciferase assay to show that the production levels were improved using this new construct.
Taken together, we described the use of the split-luciferase assay for the production of proteins by B. subtilis. We consider this assay a useful first step to screen an array of protein production conditions and select the most promising based on the luminescence levels.
We advise our colleagues who study protein secretion to add this molecular flashlight to their protein detection toolbox, either with batteries supplied or attached to the studied protein!
