Type VI secretion system: the ultimate weapon for microbial warfare
Posted on January 2, 2024 by Amy Anderson and Becca Morell
Amy Anderson and Becca Morell take us behind the scenes of their latest publication 'Distribution and diversity of type VI secretion system clusters in Enterobacter bugandensis and Enterobacter cloacae' published in Mcirobial Genomics.
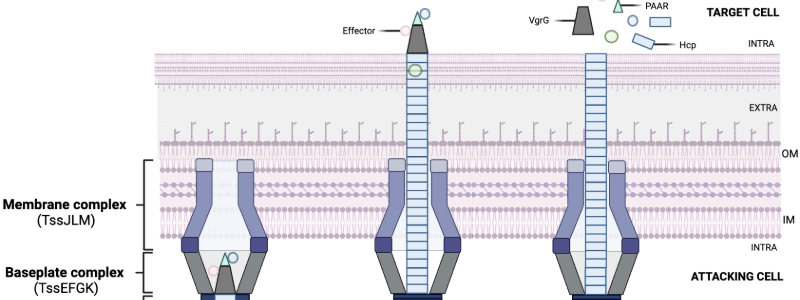
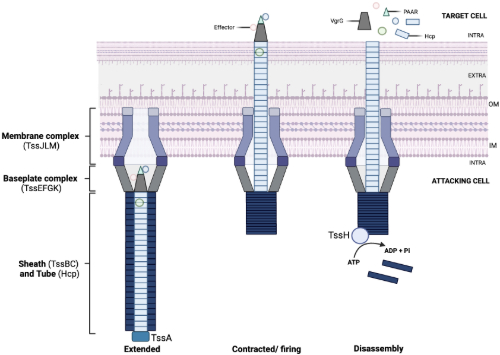
A silent war rages in every one of us as bacteria fight for survival and niche dominance within our bodies. The Type VI Secretion System (T6SS) is an excellent weapon for this arduous endeavour. It acts like a contractile molecular syringe to inject a payload of toxins (effectors) into target cells (Image 1). Bacteria not only use T6SSs to outcompete other microorganisms, but also in biofilm formation, host immune evasion, and ion scavenging.
We are Amy Anderson and Becca Morrell. Amy Anderson is a PhD student, and Becca Morrell was a work-placement student in Professor Miguel Valvano’s group at the Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Northern Ireland (Image 2). Our research group focuses on the molecular interactions of antibiotic-resistant, opportunistic bacteria with host immune cells. We investigate how these nasty pathogens establish a niche to thrive in immunocompromised individuals. Our group works on three model pathogens: Burkholderia, Achromobacter, and Enterobacter species. Amy and Becca are interested in the Enterobacter T6SS and how it can be used to manipulate bacteria and eukaryotic cells.

Enterobacter species belong to the ESKAPE pathogens. They are a group of highly multi-drug resistant and pathogenic bacteria that are the leading cause of hospital infections. The WHO listed these bacteria as requiring urgent development of new antibiotics. Enterobacter species can cause a range of infections including bacteraemia, pneumonia, meningitis, intestinal infections, cerebral abscesses, urinary tract infections and post-surgical wound infections. Although not all species are pathogenic, some have emerged as important pathogens and have been responsible for multiple sepsis outbreaks in hospitals. These include E. cloacae and E. bugandensis. E. cloacae is currently the most frequently isolated Enterobacter species while E. bugandensis is an emerging pathogen and believed to be the most virulent species within the genus.
One question that remains unanswered is how multi-drug resistant Enterobacter species colonise the gut and whether the T6SS is relevant. Given the diversity of species within the genus and its evolving taxonomy, we wanted to first characterise the distribution and diversity of T6SS components. We focussed on the clinically relevant species, E. cloacae and E. bugandensis. First, we assembled a curated dataset of 31 strains and demonstrated that most of them encode a highly conserved T6SS cluster, and 1-or-2 additional clusters that were less conserved. Next, we focused on a pathogenic E. bugandensis clinical isolate for comprehensive in silico effector prediction, with comparative analyses across the 31 isolates. Several new effector candidates were identified, including an evolved VgrG with metallopeptidase domain and Tse6-like protein. We also discovered an anti-eukaryotic catalase (KatN), M23 peptidase, PAAR, and VgrG proteins. Our findings highlight the diversity of Enterobacter T6SSs and uncover new putative effectors that may be important for the interaction of these species with neighbouring cells and their environment. This diversity may help these bacteria to establish a niche in the gut, and other mucosal tissues.
We did not have much experience in bioinformatics and had to learn from scratch. In fact, Becca discovered her flare for bioinformatics during this project and now wants to pursue it as a career! We also collaborated with Dr. Guillermo Lopez Campos - a health bioinformatician in our centre - who critically examined our results and workflow. During this study, we developed a framework that can be extended to other Enterobacter species and T6SS-weilding bacteria. In fact, we are now creating a more formal pipeline. We hope other researchers can use our framework for in silico T6SS analysis, and that the effector candidates we found can provide new avenues to elucidate the poorly understood infection biology of Enterobacter species.
‘Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else, you must run at least twice as fast as that’ – Red Queen, through the looking-glass (Lewis Carroll). Like Alice and the Red Queen, we are in an arms-race against bacteria to develop novel treatment strategies to overcome these pathogens. Other ESKAPE bacteria are known to use the T6SS for pathogenesis and bacterial competition; it is important we study its role in the Enterobacter pathogenesis scheme. Also, elucidating the mechanism-of-action of anti-bacterial T6SS effectors may provide novel strategies for antibiotic development. As we look to the future of T6SS research, studies demonstrate potential for the T6SS in vaccine development, and as a drug target to treat bacterial infections. Afterall, ‘you can learn from anyone, even your enemy’ – Ovid.