Cracking the genetic code of our cities
08 November 2016

Researchers around the world aim to map the urban genome
It is rare to say that one has lived through a revolution, but we are all living through one right now. High- throughput sequencing technologies have become cheaper and more cost-effective over the past decade, moving even faster than Moore’s Law for computer power (doubling every 18 months). Because sequencers are modern-day 'molecular microscopes', scientists believe that we are currently experiencing a scientific revolution similar to the one sparked by Antonie van Leeuwenhoek’s invention of the world’s first light microscope in the 17th century.
The past few decades have witnessed a surge in microbiome and metagenomics studies, all of which are intent on elucidating the vast invisible world beneath our fingertips. With the advent of next-generation sequencing technologies, we are able to study this world like never before.
The birth of PathoMap
In 2010, Dr Christopher Mason, Associate Professor in the Department of Physiology and Biophysics at Weill Cornell Medicine, was suddenly struck by inspiration as he watched his daughter play with toys with her friends on the train. The infants unknowingly exchanged bacteria with one another as they drooled on their toys, passing them from hand to hand, and rolling them on the ground before repeating the cycle. These observations eventually gave birth to the PathoMap study in 2013, in which Dr Mason and his research team created a molecular portrait of the New York City subway system. Inspired by Dr Mason’s daughter, the PathoMap study aimed to pioneer the field of metagenomics (study of all kingdoms’ environmental DNA) and microbiome studies by creating the first baseline geospatial metagenomic map of a city’s mass-transit system.
The PathoMap study’s primary objective was to study the microbiome of a metropolitan environment. Since New York City’s subway is the most highly trafficked transit system in the United States, it made perfect sense to sample all 468 stations to investigate the genetic and microbial diversity present in each station. A team of volunteers collected and processed a total of 1,547 samples. Using next-generation sequencing technologies and bioinformatics to analyse the sequencing data, the research team was able to determine the taxonomic classification and functional diversity of the micro-organisms present on such ubiquitous surfaces as handrails, wooden benches, train seats, rubbish bins and floors. Most of the DNA uncovered from these surfaces matched bacteria associated with the skin microbiome. Altogether, the researchers found over 600 species of microbes riding the subway with fellow New Yorkers. Furthermore, while the research team found evidence of antibiotic resistance markers and antibiotic resistant microbes, there was no evidence of virulence factors or pathogenicity in the samples. Interestingly, nearly half (48%) the DNA matched no known organism, which is a testament to the fact that we are only as good as our databases. As the field develops, our databases will become more refined and replete with accurate references that may impact the finality of taxonomic classification at species- and strain-levels.
PathoMap’s implications for human health, illness and disease
Creating the world’s first-ever metagenomics profile at the city-scale has tremendous implications for the future of public health and epidemiology. For one thing, it is the first step to creating futuristic real-time pathogen monitoring systems in urban spaces to prevent the rapid spread of epidemic and pandemic-scale disease outbreaks. The study may also help urban planners and engineers better utilise microbial ecology to design sustainable and healthy cities. Specifically, this study opens the door to incorporating the microbial world in our understanding of how building materials complement the urban microbiome. We are unknowingly building urban microbiomes each time we construct a new building or renovate a space. Studies like the PathoMap study suggest that it is crucial to incorporate a comprehensive understanding of the microbiome in order to improve upon environmental and human health. The research team was able to discover bacteria that digest toxic sludge, which may potentially help city planners and researchers formulate sustainable methods of revitalising ‘Superfund’ sites like the Gowanus Canal in Brooklyn, New York.
The PathoMap study also provides a unique model of data collection and processing to the emerging field of participatory disease surveillance, whereby community members themselves report on illnesses that emerge in close proximity to them. The use of citizen science and crowdsourcing models has further closed the gap between science and society. Dr Mason and his colleagues believe that PathoMap is a testament to the power and potential of publically engaged scientific initiatives.
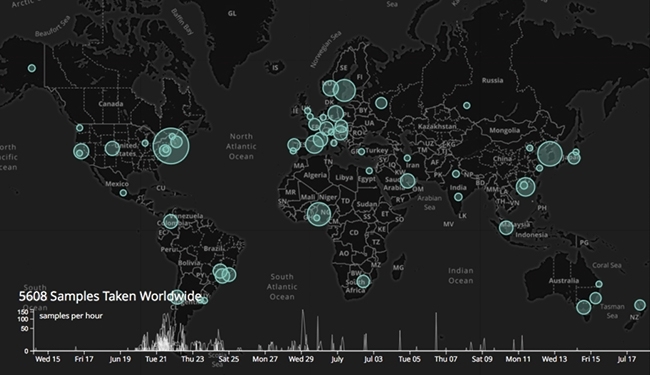
METASUB GLOBAL SAMPLE COLLECTION MAP
Inauguration of the MetaSUB International Consortium
The success of PathoMap led to the expansion of the project to other cities like Buenos Aires, Argentina; Tokyo, Japan; Cairo, Egypt; Lima, Peru; and Paris, France. In 2015, Dr Mason inaugurated the Metagenomics and Metadesign of Subways and Urban Biomes (MetaSUB) International Consortium in an effort to create the world’s first-ever longitudinal metagenomics profile of cities around the world. Since then, it has grown to include over 58 cities across 32 countries. Scott Tighe, one of MetaSUB’s premier contributing members on developing extraction techniques for the consortium, said, “The scientific reward of benchmarking the microbial DNA content of global urban environments is an awesome undertaking.” Different cities are profiling environments other than those found in the transit systems – Vienna has sampled the Danube Canal, Montevideo in Uruguay has sampled the city’s beaches (MetaBEA) and sewage (MetaSEW) systems, and Tokyo plans to sample the city and university buildings. Gaston Gonnet, the Principal Investigator of MetaSEW and MetaBEA in Montevideo, remarks that “these types of projects are quite novel and the possibility of exchanging information is very beneficial; it gives us new ideas and saves time and false starts.”
The MetaSUB Consortium has so far hosted two international meetings – one in New York City, USA, and the second in Shanghai, China – where collaborators discussed the latest updates in metagenomics research and standardised experimental protocols. Moreover, consortium members participated in ‘Global City Sampling Day (CSD)’ on 21 June 2016 in concert with Ocean Sampling Day to map genetic and epigenetic stratification of antimicrobial resistance markers in the urban environment. Dr Leming Shi, co- organiser of the 2nd Annual MetaSUB Summit in Shanghai, commented on MetaSUB’s unique model of scientific collaboration. He said, “What impressed me the most is MetaSUB’s capability of engaging a mix of well-accomplished scientists, young college students, and graduate students under the same roof with the same objective of gaining a better understanding of ourselves by better understanding the environment we live in.”
GEOSPATIAL MAP OF ENTEROBACTERIACEAE
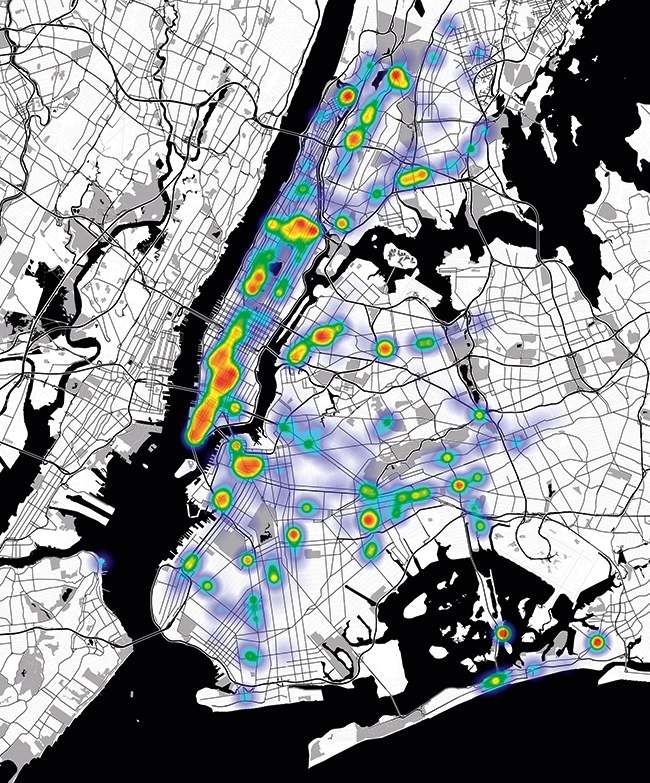
Mapping human ancestral data in MetaSUB Sheffield, United Kingdom
In Sheffield, UK, Dr Eran Elhaik is working hard to piece together human ancestry data from metagenomic data found on public surfaces. Elhaik states:
“One of the greatest difficulties in studying microbial ecology is their complex relationships with human populations. Inferring the geographical origins of humans from the genetic data collated during the swabbing process allows identification of the demographic forces that shaped the microbial communities. However, despite its great promise, due to technological difficulties, we are only capable of studying the combined DNA of individuals or rather ‘communal DNA.’ Afshinnekoo & others already showed that when classifying this ‘communal DNA’ to four ethnicities, their proportions are correlated with the ethnicities in the Census data. We extended this approach by classifying the ‘communal DNA’ to twelve potential gene pools and applying the GPS algorithm, shown to infer geographical origins with high sensitivity (0.75) and specificity (0.99) (Elhaik et al. 2014). Compacting the 12 gene pools to four and applying GPS to the NYC subway data yields results that are in agreement with those of Afshinnekoo & others and the Census data. Our approach can thereby be applied to infer temporal population dynamics and study their effect on micro-organism communities.”
Exploring microbial diversity in MetaSUB Sydney, Australia
Rather than focusing on human ancestral data, Dr Aaron Darling and Dr Catherine Burke of the University of Technology Sydney, Australia, are focusing their efforts on delineating the taxonomic classification and functional diversity of microbes in the Sydney transit system. Dr Burke emphasises that her interests lie in exploring the effect of natural sources of air ventilation and exposure on microbial diversity in built environments because they are further correlated with positive health outcomes, like a decreased risk of asthma. Because Sydney’s city train stations are exposed to a variety of different environments, she and Dr Darling are interested in seeing the effect of these different exposures on the microbiome of each station, such as outdoors vs underground, and harbour vs further inland train stations. She states, “Knowing how exposure to different environments affects microbial diversity in these public spaces could help inform their future design.”
The 2016 Olympiome
Along with the launch of Global CSD, MetaSUB is launching the world’s first ‘Olympiome’. Co-organised by Drs Emmanuel Dias-Neto, Milton Ozório Moraes, Fernanda Kehdy, and Christopher Mason, the researchers profiled Rio’s subway and other public areas before, during, and after the 2016 Rio Olympics. This initiative will better reveal how migration at large-scale public events impacts the microbiome and elucidate the genetic signatures that move between cities. Dr Dias-Neto said of the Olympiome project that it is “a very exciting project” because it is the “first time the microbiome will be studied in a big global mass event.”
SOFIA AHSANUDDIN, EBRAHIM AFSHINNEKOO & CHRISTOPHER E. MASON
Department of Physiology and Biophysics, Weill Cornell Medicine, NY, USA; The HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Weill Cornell Medicine, NY, USA
[email protected]
[email protected]
[email protected]
FURTHER READING
Afshinnekoo E. & others (2015). Geospatial Resolution of Human and Bacterial Diversity from City-scale Metagenomics. Cell Systems 29 July 2015.
The MetaSUB Consortium (2016). The Metagenomics and Metadesign of the Subways and Urban Biomes (MetaSUB) International Consortium Inaugural Meeting Report. Microbiome 3 June 2016.
