Fluorescence in situ hybridisation: making microbes glow to see how they organise
13 February 2018
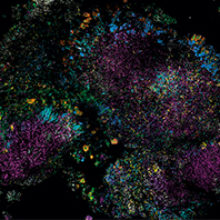
The microbial world is social, but we usually study it in monoculture. Most of what we know about the behaviour and properties of microbes comes from studying pure cultures, single cells allowed to proliferate into millions in the laboratory in sterile broth or agar.
But in the natural world, a microbe lives in a complex and variable environment. Its neighbours can cause it to change its metabolic profile, and a microbe embedded in a cluster of other microbes can have properties that it would not have on its own, such as the ability to survive in an aerobic environment or invade a host cell. If we want to understand the interplay of human and microbial in our own biology, or if we want to understand how microbes degrade toxins, help plants grow or drive biogeochemical cycles in the ocean, we have to understand how microbes behave within communities.
Micron-scale spatial organisation is key to understanding microbial community behaviour. Bacteria are micron-sized and frequently occupy habitats that are spatially heterogeneous at micron scales. Important features of the environment, from a microbe’s point of view, can include inorganic material, plant or animal tissue, organic detritus, and the mucus and mucilaginous goo secreted by animals and plants, as well as other bacteria, all of which can provide surfaces for attachment and sources of nutrients. Understanding the structure of these habitats, and the localisation of microbes within the habitat at micron scales, is a first step in elucidating how the communities work.
Glowing reporters
Microscopy is a uniquely suitable technique for looking at microbial communities and learning about their structure and organisation. But with hundreds of types of bacteria in any given sample, and with only a few visible morphologies, such as spheres, rods, and filaments, many disparate bacteria look near-identical under a microscope.
This identification problem can be solved using fluorescence in situ hybridisation (FISH). FISH is a technique in which a probe made of DNA or RNA is used to locate a complementary DNA or RNA molecule in a cell by binding (hybridising) to it. Each molecule of probe is attached to a small fluorescent tag that can be detected by microscopy – a glowing reporter molecule that reveals the presence and the location of the target.
In medicine, FISH is commonly used as a diagnostic test for chromosomal abnormalities. Microbiologists use it in a different way – to visualise where the bacteria are in a sample, and also to identify bacteria using probes complementary to the ribosomal RNA of only certain groups of bacteria. FISH lets us track the subtle changes in localisation of bacteria, showing exactly where they are within the tissue of a host, for example, and how that localisation changes when we change some feature of the host.
FISH is instrumental in visualising individual microbial types and where they are. But for visualising interactions in natural communities, there is a mismatch between the complexity of the community and the number of different types that traditional FISH can see. FISH is usually done using a simple fluorescence microscope with bandpass filters that can differentiate three or four bands of emitted fluorescence – such as far-red, red, green, and blue wavelengths. For an experiment with several probes, the fluorophores for each probe have to be chosen so that their fluorescence emission spectra overlap as little as possible, so that each can be discriminated by a different filter. This requirement for minimal overlap usually limits to three or four the number of signals that can be simultaneously imaged.
Hyperspectral imaging expands the palette
Hyperspectral imaging is a technology that leapfrogs the limitations of bandpass filters and brings the capabilities of FISH much closer to matching the complexities of a natural microbial community. With hyperspectral imaging, it is possible to read the fluorescence spectrum from every pixel in an image and differentiate fluorophore signals that overlap spectrally – making it possible to discriminate a dozen or more fluorophores. Using this technology, my colleagues, including Gary Borisy, Floyd Dewhirst and Alex Valm, and I developed a way to tag and image most or all of the major members of a microbial community simultaneously. We called it CLASI-FISH: combinatorial labelling and spectral imaging FISH, and with it, suddenly the structure of complex microbial communities, the spatial relationships of the bacteria to one another, became visible.
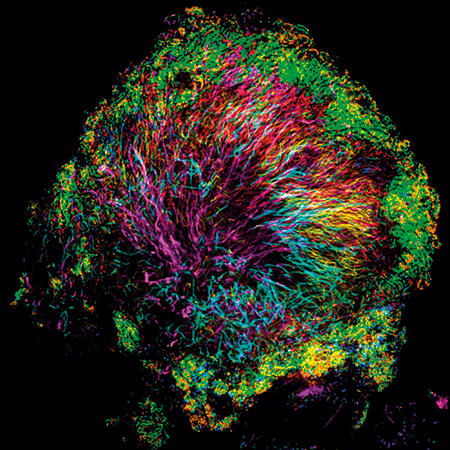
Amazing structure
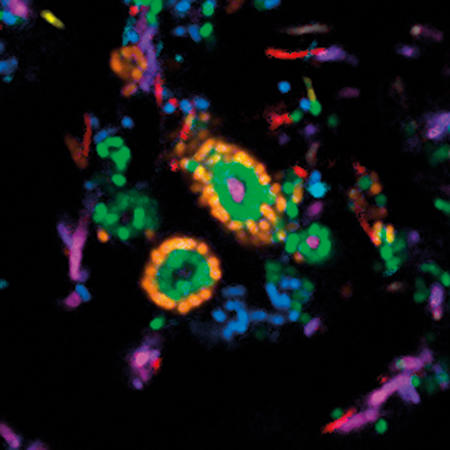
Using CLASI-FISH, we found an amazing microbial structure very close to home – on the surface of teeth. We scraped plaque from the teeth of volunteers, dropped it onto a slide, applied a set of probes, and saw an astonishingly complex and highly organised microbial community. Filaments of the microbe Corynebacterium clustered together and radiated outward, forming a clump or bush whose sometimes spiny appearance suggested the name we gave to this community: the hedgehog. The other microbes making up the hedgehog structure inhabited the Corynebacterium bush at characteristic positions. Oxygen-tolerant taxa such as Streptococcus bound to the tips of the filaments and formed a kind of outer shell, and anaerobes sheltered in a presumably low-oxygen band just inside this shell.
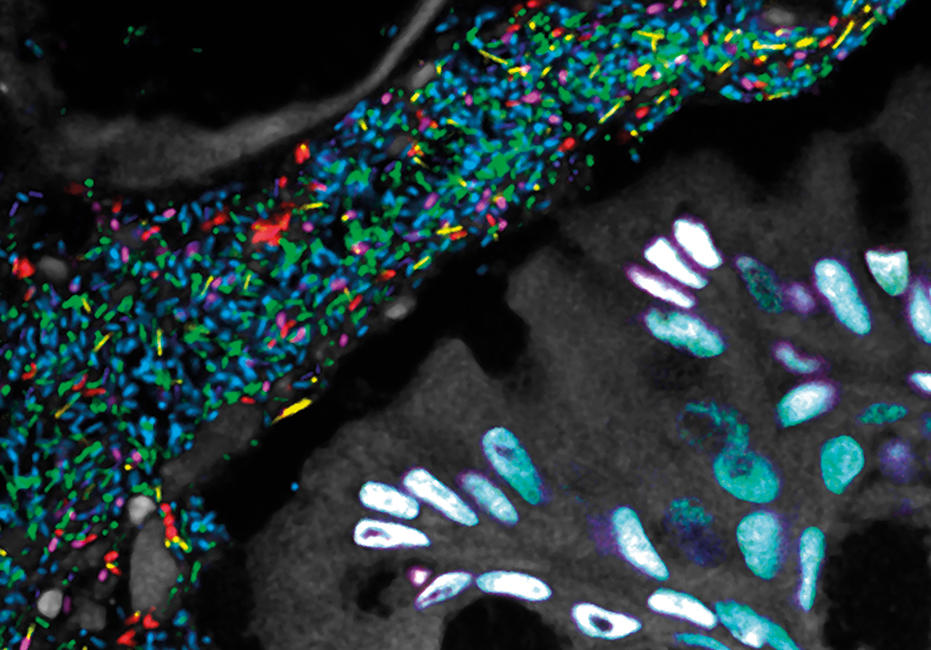
As we had hoped, seeing the structure immediately suggested hypotheses about function and the inter-relationships of taxa: which taxa form the physical scaffold on which the community is built; which appear to play a key role in modulating the biochemical environment; and which are localised in a strikingly specific way adjacent to one another, their physical closeness suggesting that they share an important biochemical interaction or a similar niche. But not every microbial community is organised in this way. Another habitat close to home – the gut – housed a microbiota with a structure that proved to be strikingly different, with less apparent spatial organisation and more micron-scale mixing.
Further reading
Langer-Safer, P. R. & others (1982). Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci U S A 79, 4381–4385.
Mark Welch, J. & others (2016). Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A 113, E791–E800.
Mark Welch, J. & others (2017). Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A 114, E9105–E9114.
Propheter, D. & Hooper, L. (2015). Bacteria come into focus: new tools for visualizing the microbiota. Cell Host & Microbe 18, 392–394.

Jessica Mark Welch
Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, 7 MBL Street, Woods Hole, MA 02543, USA
Jessica Mark Welch is an Associate Scientist at the Marine Biological Laboratory in Woods Hole, Massachusetts. In addition to her work as a scientist, she serves as host to a varied and structurally complex community of microbes.
Does seeing the structure of these microbial communities change how we think about them?
We know that different kinds of plaque coexist in the mouth and have different properties – for example, that plaque rich in filaments is more likely to form dental calculus than plaque rich in cocci. But whether the presence of either type of plaque makes the preservation of health, or a possible transition to disease, more likely we do not yet know. FISH and imaging, however, can sharpen our questions, focus our attention, and point out promising avenues for targeted investigation to define the role of the microbiome and its different conformations in healthy people.
What next?
I hope that the hedgehog is only the first in a long line of complex spatial structures we will discover with CLASI-FISH, so that we can begin to understand the common principles underlying diverse structures. Other fluorescence microscopy techniques provide exciting complementary approaches. As well as being used to identify cells, FISH can be tuned to detect single-copy genes and gene expression, moving us closer to an understanding of how individual microbes and the community as a whole react to changing conditions. Live imaging at high resolution provides an exciting window into community dynamics. All of these methods contribute toward the goal of being able to understand, predict, model, and manipulate the function of these complex and beautiful microbial communities.
Images: The first, thumbnail image shows how bacteria form distinctive, organised structures in human dental plaque. The second depicts the radial organisation of bacteria in a 'hedgehog' structure from dental plaque. The third image demonstrates the mixed organisation of bacteria in the mouse colon, and the final image shows the 'corncob' structures from dental plaque. All images by Jessica March Welch.
