Adaptation and Amino Acids: How Experimental Evolution Revealed a New Pathway for Staphylococcus aureus Tolerance to Pseudomonas aeruginosa
Hello, my name is Dr Ashley Alexander, I am a postdoctoral researcher at Georgia State University, Atlanta, Georgia, USA, in Dr Sheyda Azimi’s research group. The work in this article was completed during my time as a PhD student in the Population Biology Ecology and Evolution program under the mentorship of Dr Joanna Goldberg and Dr Tim Read at Emory University, Atlanta, Georgia, USA.
Experimental evolution is a tool that can be applied to many systems, including microbial interactions. As a new graduate student, I was excited to use this method to investigate the question: “how will Staphylococcus aureus adapt to repeated exposure to another species that effectively inhibits its growth, like Pseudomonas aeruginosa?”. Taking the perspective of an evolving population of S. aureus — I could view P. aeruginosa as similar to an antibiotic, or any factor that might exert strong selective pressure on each new generation. Using a method of repeatedly coculturing S. aureus and P. aeruginosa together, I intended to watch and learn as S. aureus evolved its way around the challenge of a problematic neighbour like P. aeruginosa.
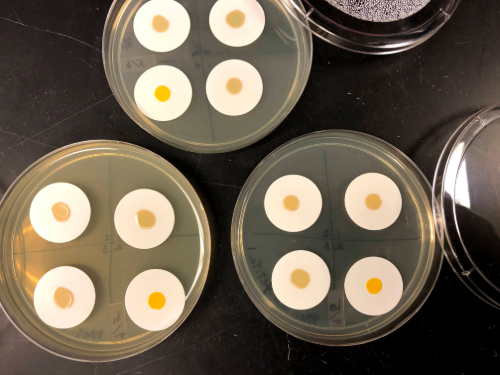
In the design of the evolution experiment, I also thought about where and how these pathogens may encounter each other in the natural world. In the case of lung infections in individuals with the genetic disease, cystic fibrosis, S. aureus tends to arrive on the scene first. When P. aeruginosa is introduced into the infection S. aureus is either replaced, adapts to survive alongside P. aeruginosa, or it continues to persist in parts of the lung where P. aeruginosa hasn’t colonised. To replicate this pattern of P. aeruginosa invading established S. aureus populations, I started each coculture with 30 times more S. aureus than P. aeruginosa. With this mixture of lots of S. aureus and small amounts of P. aeruginosa, I inoculated cocultures on solid agar and used filter paper to keep the culture contained and easy to recover. After each coculture period, I would recover the surviving S. aureus and use them to start the next passage - hopefully selecting for S. aureus that were better at coexisting with P. aeruginosa. I repeated these consecutive cocultures 8 times and found that out of my four replicate populations, two had become tolerant to P. aeruginosa - maintaining most of their population size whenever they were cocultured with P. aeruginosa, compared to the original S. aureus where very little S. aureus survived in the presence of P. aeruginosa. This finding was exciting, but the best was yet to come — when we looked at the genomes of isolates from one of the Pseudomonas-tolerant populations we found that there was a single site of disruptive mutations that were unique among the tolerant isolates in an amino acid transporter, gltT. The mutations in gltT actually occurred in two different ways in the same population — meaning there were two isolates with similar but unique mutations. This implied that disrupting this gene was highly selected for during the evolution experiment. With this observation in hand, we then set out to determine what gltT did for S. aureus in the presence of P. aeruginosa and why S. aureus had repeatedly disrupted this gene in this experiment. We analysed the role of gltT in varying nutrient conditions, in in-vivo mouse infections, amino acid uptake and looked for growth differences among wild-type and mutant strains microscopically. The evidence that we found from our analysis has led us to the postulate that disrupting gltT allowed S. aureus to gain Pseudomonas-tolerance by balancing its intake and metabolism of key amino acids. This may be especially relevant in our laboratory setting of a nutrient rich environment and P. aeruginosa’s effectiveness at consuming large amounts of amino acids. Interestingly, gltT is highly conserved in nature — among over 444 representative genomes we did not find any mutations that were similar in their function to what we found in our evolution experiment.
There are many exciting future directions for this work, including testing this experimental evolution approach with clinical isolates and further investigation of how S. aureus changes its nutrient uptake and metabolism in the presence of other CF pathogens like Stenotrophomonas maltophilia. As for how this work may contribute to our understanding of cystic fibrosis related infections — experimental evolution showed us that there are many mechanisms of interactions and tolerance between S. aureus and P. aeruginosa that we don’t fully understand yet. Factors like metabolism and nutrient uptake may be overlooked and are important traits to consider when we study and work to treat polymicrobial chronic infections.
I am excited to continue to contribute to the research on microbial interactions between opportunistic pathogens like Staphylococcus aureus and Pseudomonas aeruginosa through the lens of their ecology and evolution because as we gain a greater understanding for how these highly adaptive organisms change over time we’ll not only learn how to better address infections by them, but we’ll also learn more about the potential that organisms have to adapt to their environments in the face of selective pressures. We may be able to extend findings from this system to different kinds of microbial communities and ecological communities in general. Microbes have been adapting to life on this planet longer than any other form of life on earth and there is so much we can learn from them just by watching how they change over time.
