The resistance: how E. coli learns to fight our most potent antibiotics
Antibiotics are our best defence against infections caused by bacteria. They have saved millions of lives since their discovery in the first half of the 20th century, but the rise of antibiotic resistance is threatening their usefulness and with it the safety of modern medicine.
At the department of Medical Microbiology and Infection Prevention at the Amsterdam University Medical Center, Netherlands, we study the mechanisms of antibiotic resistance in bacteria recovered from patients in an effort to improve our methods of detection and prevention. Early detection of antibiotic resistance determinants means we can prescribe the correct antibiotic to a patient immediately without trial and error. We can also efficiently isolate patients at risk and reduce the risk of transmission of resistance to other patients.
Bacteria are constantly evolving and use this to their advantage to devise new ways to resist our most potent drugs. They can acquire new genes that code for specific enzymes that target and degrade the antibiotic molecules. They can also modify the proteins that regulate the flow of molecules through the cellular membrane, called porins, in an effort to prevent the antibiotics from reaching their target within the cell.
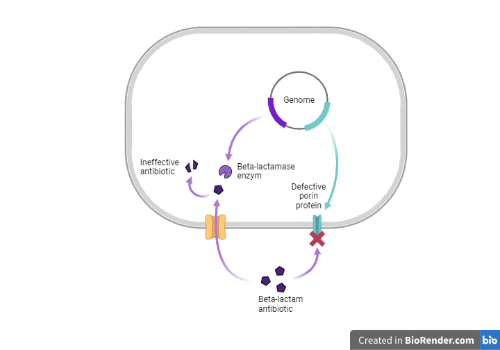
Escherichia coli can cause bloodstream infections in vulnerable patients, a life-threatening condition treated by the administration of antibiotics such as meropenem, a very broad-spectrum beta-lactam antibiotic that belongs to the carbapenem class. E. coli can become resistant to carbapenems by acquiring genes called carbapenemase, which are more powerful versions of the traditional beta-lactamase ones. In vitro experiments have shown that E. coli can become also resistant to carbapenems (including meropenem) by modifying the sequence of the ompC gene: the gene then produces a defective version of the OmpC porin protein, greatly reducing its capacity to transport antibiotics inside the bacterial cell.
A patient in our hospital was diagnosed with an E. coli bloodstream infection. Initially, we recovered a meropenem susceptible E. coli isolate from the blood of this patient. Eight weeks later, the same E. coli clone was isolated from the patient, but this time showed meropenem resistance. None of our tests could detect the cause of this resistance, the acquisition of a new carbapenemase gene for example. Even sequencing the complete genome of the two E. coli clones could not give a definite answer. We speculated that, because the Illumina technology we used for sequencing is not adapted to detect insertions, deletions or rearrangements of genes, we were looking at an incomplete picture. In addition, we decided to also use Nanopore technology to produce long DNA fragments, which can be combined with Illumina ones to produce high quality complete genomes. Indeed, comparing the two completed genomes showed some notable differences.
First, a CTX-M-15 extended-spectrum beta-lactamase gene present as a single copy in the genome of the first isolate was found three times in the second isolate. This could have led to production of higher amounts of the enzyme in an effort to try to degrade antibiotics more efficiently. We then determined that the ompC gene in the second isolate was modified by the insertion of a 1200 bp sequence. This type of insertion can lead to production of defective OmpC porin proteins, but its acquisition in vivo in a patient during treatment is a new finding.
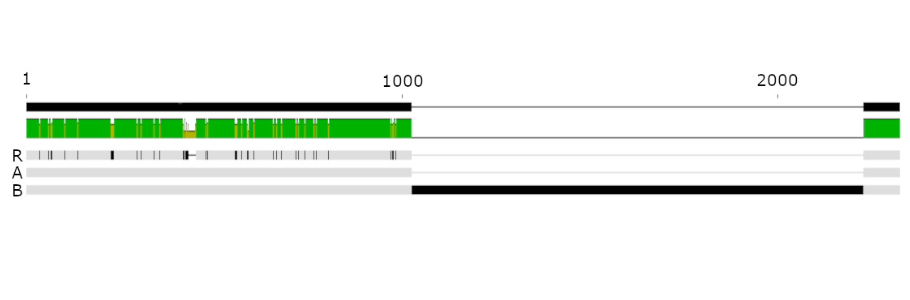
Antimicrobial resistant bacteria represent a serious threat to the health of fragile patients, potentially leading to very heavy antibiotic regimen or even untreatable infections. In the present case, the resistance arose in less than 8 weeks while the patient was under treatment. Common microbiology and molecular methods could not detect the origin of the resistance. By sequencing the complete genome using simultaneously short (Illumina) and long (Nanopore) reads technology we could elucidate the potential responsible mechanism. We are now working on implementing this technology for our routine diagnostic, providing even more accurate diagnostic for our patients, and optimizing each treatment.
Thumbnail image created with created with BioRender.com
