Democratising Bioinformatics! Breaking the Bioinformatics Barrier in AMR Genome Analysis
Are you a mid-career antimicrobial resistance (AMR) wet-lab researcher by training, who (i) found yourself suddenly crash-landing in the era of genome sequencing, (ii) do not have any bioinformatics training and (iii) would like to break the bioinformatics barrier in AMR genome analysis without wrangling with coding?
Our story started with the opening of a subgrant for AMR funded by the Public Health Alliance for Genomic Epidemiology (PHA4GE) in early 2021. The subgrant called for teams to test and implement hAMRonization, a standardized AMR gene detection pipeline developed by PHA4GE for analysis and comparison of AMR gene content from genome sequences.

We are a team comprising of four bioinformaticians (Su Datt, Sabrina, Mia Yang, Emma – our mentor from PHA4GE), one infection control specialist (Zetty) and two microbiologists (Sheila and Hui-min), banded together and field-tested hAMRonization on methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from Malaysia and Argentina (for those asking why these 2 countries? Well, most of us in the team are from Malaysia, and Hui-min got connected with Sabrina from the Train the Trainer: Design Genomics and Bioinformatics Training course).
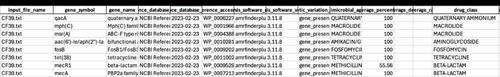
From our testing, we found that even though the MRSAs were from two very different regions of the world, as long as genome sequences were available, AMR gene content from the strains could be compared between laboratories. The hAMRonization results output are in the form of a spreadsheet (see above), AMR analysis results using tools (such as AMRFinderPlus, ResFinder) and epidemiological data of tested strains are listed in columns.
However, we noticed that coding skills are still required to operate hAMRonization. As (i) PHA4GE intended for the pipeline to be useful for public health specialists, and (ii) knowing the #pain experienced by wet-lab biologists who have no bioinformatics training in performing genome data analysis, we thought of going one step further to “democratise” the analysis process via compilation of AMR analysis tools and hAMRonization into a Google Colaboratory Notebook, which we coined as AMRColab. We proceeded to hold an online workshop (see above) in 2022 to publicize the utility of AMRColab. Workshop participants were mostly Malaysian public health specialists, and, being wet-lab personnel, they were surprised at the ease of using AMRColab for AMR gene analysis, without the need for coding! (yayy!). A manual was also developed for the Colab.

To further improve AMRColab, we thought that having some plug-ins, such as the one from Microreact, will be helpful for researchers to visualize their AMR gene comparison results (original output in the form of spreadsheets). We did this and feedback received from a second workshop in 2023 (see below) convinced us that the Colab is indeed helpful and successful in breaking the bioinformatics barrier in AMR genome analysis. Future plans on the horizon for the Colab include translation of the manual into more languages (Spanish will be first!), having a YouTube tutorial (for those who dislike reading … manuals), and perhaps an online community platform. And… if you are wondering if the Colab can now process your AMR genome sequences fresh out from the sequencer into beautiful visuals for your tested strains, ponder no more, as the beta version for this function is now ready (shoutout to reviewer number 1 of our manuscript for this wonderful idea!).
We thank PHA4GE for the opportunity to have banded together for this journey in “democratising” bioinformatics for AMR wet-lab researchers and public health specialists. Huge appreciation to our workshop participants and our manuscript reviewers who provided very constructive feedback in making a functional Colab a reality. The Colab is also being developed as part of a bigger project to investigate AMR carriage in the community – a project sponsored by the Ministry of Higher Education, Malaysia.
Moving forward, we hope all of us in the team can finally meet up (yes, some of us have never met in person except online!) We are happy to collaborate with all AMR enthusiasts and funders (contact [email protected] / [email protected]). Our dream is that the AMRColab will indeed help break the bioinformatics barrier in AMR research and surveillance, allowing the research and public health community to always, always stay ahead of the AMR curve.
Image: iStock/Sergey Shulgin
