How do your microbes grow?
Posted on December 16, 2015 by Anand Jagatia
If you’ve recently been on holiday, changed your diet, or taken antibiotics, the chances are that it’s affected your microbiome. This community of microbes – made up of trillions of bacteria, archaea and fungi – plays a vital role in our health, but it’s also sensitive to changes in our external and internal environment.
This is why it’s important to see how these micro-organisms behave in places like the gut, if we hope to understand how they might be linked to things like allergies and mental health. The problem is that your gut is like a black box – microbes inside can’t be observed directly, so it’s difficult to measure their growth. If you were so inclined, you could analyse the microbial DNA in people’s faeces, but this only gives you an overall sense of what’s happening; it can’t tell you the growth rate of individual species in response to specific factors.
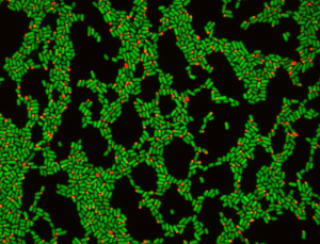
But now, researchers from the Wyss Institute at Harvard University have developed a technique that can do just that. The way it works is simple but powerful – essentially, you genetically engineer bacteria to produce a bright red particle inside themselves. This particle is fluorescent and is made up of thousands of proteins. However, it’s only produced by the first generation of bacteria – so when each of these divides in two, the particle is transmitted to just one of the resulting cells. After one generation only half the bacteria will contain a particle, after two generations a quarter, and so on.
The fraction of cells with a red dot tells you how many generations have passed.
The team used this method (called DCDC) to “see” the growth of E. coli inside a mouse gut over time. The bacteria that went into the mouse all contained a marker, but when they came out the other side only some of them did. The fraction tells you how many generations of growth have elapsed.
“The technique is great for telling us about the growth of specific strains, but these can also serve as an indicator for what’s happening in the rest of the gut.” says Cameron Myhrvold, lead author on the paper, which was published in Nature Communications. “So we can use our engineered strains as a proxy for other ones.”
The principle behind the method is actually borrowed from ecology, something called mark and recapture, where researchers physically mark animals with a tag or paint. But when it comes to very small organisms, things become more difficult.
“People haven’t previously applied this technique to cells,” says Cameron. “The smallest things people have tried is something like bees, which does work, but it’s very laborious. As the organism gets smaller, it’s harder to mark them, but there are also more individuals for you to mark. So it’s a double whammy.”
For this reason, DCDC gets the bacteria to mark themselves. Expression of the particle is switched on using a chemical promoter, and then switched off, ensuring only the first group of cells have been marked. The marker itself also has to be physically stable – otherwise, one particle breaking in half would look like two particles, and throw the numbers off. “As the cells grow exponentially, the number of cells with a particle becomes very low,” Cameron explains. “So stability was really important, as was having a low false production rate.”
DCDC could be used to accurately measure rapid changes in the microbiome, as the authors demonstrate in their paper. But in theory, it can be applied to any area of microbiology where observing the growth of individual species is hard. “One thing that caught my attention is applying this to fermentation, which often involves several different strains fermenting at the same time,” Cameron says. “It’s very hard to know how they are all growing together to produce the final product, so this technique could help us to understand and control that.”
Myhrvold, C., Kotula, J., Hicks, W., Conway, N., & Silver, P. (2015). A distributed cell division counter reveals growth dynamics in the gut microbiota Nature Communications, 6 DOI: 10.1038/ncomms10039
