M.bovis infection in cattle and badgers. Genomics brings nuance to a ‘black and white’ issue
Posted on June 7, 2023 by Dr Adrian Allen
Dr Adrian Allen takes us behind the scenes of his latest publication 'Genomic epidemiology of Mycobacterium bovis infection in sympatric badger and cattle populations in Northern Ireland' published in Microbial Genomics.
My name is Adrian Allen, I’m a Principal Scientific Officer at the Agrifood and Biosciences Institute (AFBI), Belfast, Northern Ireland (NI).
I lead the pathogen genomics team at AFBI’s Molecular Bacteriology Branch, a public laboratory whose work is undertaken for the Department of Agriculture, Environment and Rural Affairs (DAERA). While we work on a variety of important veterinary pathogens, the one which dominates our work is Mycobacterium bovis, the causative organism of bovine tuberculosis (bTB), which currently affects ~10% of cattle herds in NI. The pathogen’s broad host range hampers eradication efforts since infection of local wildlife can spill back over into cattle. In the UK and Ireland, the primary wildlife host for bTB is the European badger.

iStock/Anne Coatesy
That badgers are involved in the epidemiology of bTB is uncontroversial. Culling trials and molecular investigations have shown infections in cattle and badgers are closely linked. What, until relatively recently, has been opaque, is the relative importance of transmission pathways – cattle-to-cattle; cattle-to-badger, badger-to-cattle and badger-to-badger. Understanding which of these predominate is of interest to policy makers who run disease eradication schemes.
Previously, M. bovis molecular epidemiology relied on genotyping small numbers of low-resolution molecular markers such as variable number of tandem repeat (VNTR) loci. These could only tell us if individual animal cases were potentially linked - investigating chains of transmission was impossible. By analogy, it’s hard to review and compare two drafts of the same blog to determine if they’re similar if you’re only looking at a few words from the entire text. Much better to read all the text before reaching a conclusion about how similar or otherwise they are. Whole genome sequencing (WGS) of the M. bovis genome is like reading all the text. We can see how closely related bacterial isolates taken from cattle and badgers are and use this to build family trees (phylogenies) which can tell us something about transmission dynamics in outbreaks.
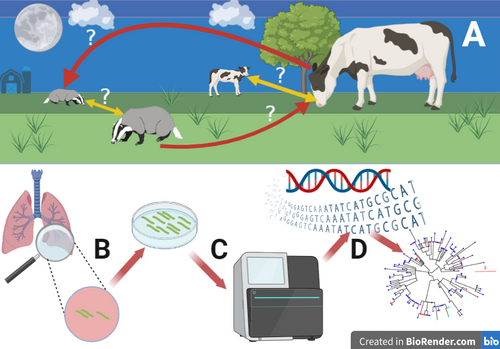
A – Cattle and badgers share the same habitat and can transmit bTB between other members of the same species (yellow arrows) and members of different species (red arrows). In the recent past, it has been very difficult to ascertain the relative importance of each transmission route. B – bTB positive badger and cattle tissue can be cultured to grow M. bovis. C – M. bovis cultures can be genome sequenced. D – Genomic data can be used to build phylogenetic trees that inform on the importance of specific transmission routes. Figure produced in www.Biorender.com
Having access to cattle and badger M. bovis isolates over a wide window of time is required to build a phylogeny for transmission dynamics inference. Fortunately, AFBI has a biobank of several thousand isolates from both species going back thirty years. Alongside this resource, DAERA undertook a selective badger culling intervention trial in a small area (100km2) of NI between 2014 and 2018 which had experienced an unexplained rise in disease incidence in 2011-2012. In this area, alongside ongoing culling of infected cattle, TB positive trapped badgers were euthanised. Tissues from both species were then sent for bacteriological culture and WGS. Using historic and contemporary isolates, this area was the perfect test bed for exploring the benefits of WGS based epidemiology.
We collected ~600 M. bovis isolates for WGS, spanning a period of 30 years, making this 100km2 area one of the most comprehensively sampled and best studied bTB breakdowns in the UK and Ireland. Our M. bovis phylogeny had eight distinct branches, seven of which contained bacterial strains that came from other regions of NI. These seven lineages were likely imported into the study area through the movement of infected cattle given that their own home ranges were found in places further away than could be facilitated by the movement of badgers.
The eighth branch in the phylogeny, was the endemic M. bovis lineage, containing most isolates. All the sub-branches coming off this major lineage had closely related isolates from both cattle and badgers which was good evidence that both host species were sharing the same infections.
By using Bayesian phylogenetic methods, we time stamped the endemic lineage, determining the ‘clock’ rate of how the DNA sequence of the bacteria changed over time. From this we determined the endemic lineage had been in the study area for 40-50 years and that around 2011-2012, the genetic diversity of this lineage had greatly increased, in keeping with the observed rise in disease incidence - a gratifying congruence of genomic and traditional epidemiological data.
Bayesian phylogenetics also permitted us to use two different approaches to assess the transmission dynamics of the endemic lineage. Both suggested cattle-to-cattle transmission was the most common route of disease dissemination while also detecting inter-species transmission. One method suggested cattle-to-badger was considerably more common than badger-to-cattle transmission, while both methods suggested badger-to-badger transmission was rare.

Uniquely, for this type of study on bTB, we also DNA profiled the badger population from the study area to determine their population structure and found that, as with other badger populations in Britain and Ireland, they exhibited ‘isolation by distance’, a feature of territorial species wherein closely related badgers live physically closer in the landscape and more distantly related animals live further apart. We hypothesised that if badgers were playing a large role in spreading disease around the study area, one might expect to observe that the most closely related badgers shared closely related M. bovis isolates. They did not.
The evidence, therefore, suggested that in this area of NI, cattle played a major role in disease dissemination, with badgers being involved, but to a lesser extent. These transmission routes do not occur in isolation from one another, however. Even a ‘trickle’ of infection from badgers to cattle could be hugely amplified by the large amount of cattle-to-cattle transmission we observed.
Our results agreed with previous studies from Britain which found intra-species transmission was the most common route of disease dissemination. However, unlike our study, in some of these locales, inter-species transmission was dominated by the badger-to-cattle route. It seems likely there is no single paradigm for bTB transmission, and that it varies by location. Accounting for such heterogeneity could help policy makers tailor interventions leading to more efficient disease eradication outcomes.
This project enhanced our existing collaborations with colleagues at the Universities of Edinburgh and Glasgow. It was also central to the ‘modernising microbiology’ agenda AFBI and many other public laboratories have been progressing in recent years at a time of unprecedented pressure on public finances – allowing us to embed routine genomic methods in house, improve capacity and develop expertise essential for maintaining biosecurity and improving animal and public health.
