Not all E. coli are equal
Posted on April 27, 2023 by Dr Hannah Pugh
Dr Hannah Pugh takes us behind the scenes of her latest publication 'E. coli ST11 (O157:H7) does not encode a functional AcrF efflux pump' published in Microbiology.
My name is Dr Hannah Pugh, I was a PhD student in the group of Dr Jessica Blair at the Institute of Microbiology and Infection (IMI), University of Birmingham, UK. I am currently a postdoctoral research fellow in the Systems Chemical Biology of Infection and Resistance laboratory at the Francis Crick Institute, UK. The work of Dr. Blair’s group focuses on the molecular mechanisms of antibiotic resistance, especially efflux pumps of the resistance nodulation division (RND) family. These are protein machines that pump a wide range of molecules from inside the bacterium out into the extracellular environment.
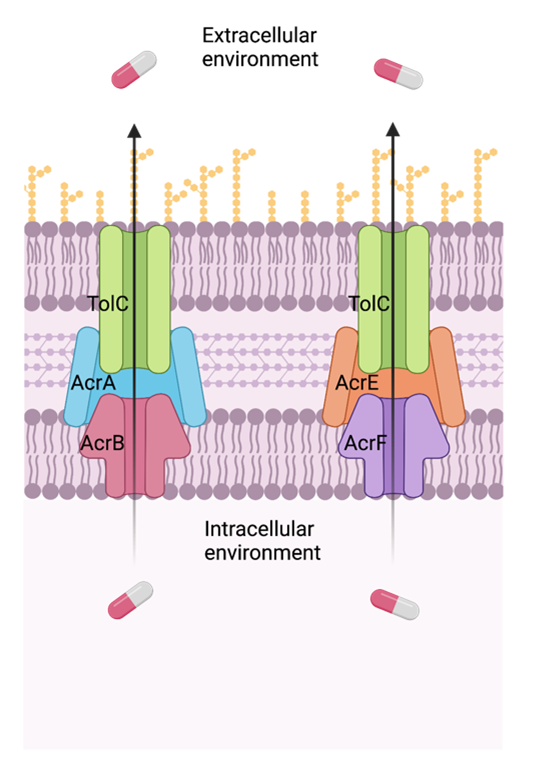
How RND pumps work (created with Biorender.com).
Escherichia coli is a bacterium that can be found throughout the environment and within the human gut, and in some cases E. coli can cause disease. Much of the work done to characterise RND pumps in E. coli has been done in a single strain, E. coli K-12 substr. MG1655, with results from this one strain frequently taken as a representative of the whole species. My PhD work asked questions about the prevalence and conservation of RND efflux pumps across a range of E. coli sequences and aimed to determine whether these assumptions were justified.
RND pumps have a wide range of substrates, from host metabolites to clinically relevant antimicrobials. Through mutations in the efflux pump or the factors that regulate them, bacteria such as a E. coli can survive higher concentrations of antimicrobials. As a result, RND pumps play a critical role in today’s antimicrobial resistance crisis.
In recent years it has become clear that there is high variation across E. coli and that no strain can be taken as a true representative of the species. With this in mind, we wanted to look at the conservation of RND pumps across E. coli. In collaboration with Dr Chris Connor, who was a PhD student with Professor Alan McNally at the IMI, we utilised the vast number of E. coli assemblies available on open-access repositories. We focused on two efflux pumps systems in particular; the genes that encode the RND pumps AcrAB and AcrEF. We also looked at TolC, as this protein forms a complex with both AcrAB and AcrEF, in addition to other E. coli transporters.
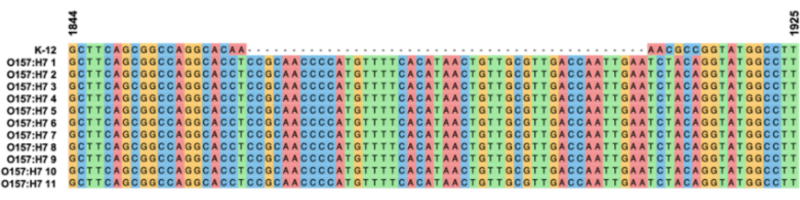
AcrAB-TolC is often thought of as the “main” RND pump in both E. coli and Salmonella enterica as it is constitutively expressed and has a very broad substrate range. We found that most E. coli types only had a single gene variant suggesting high conservation of the AcrAB-TolC system. AcrEF has a very similar substrate range to AcrAB but unlike AcrAB, the genes that encode AcrEF are not expressed in the laboratory. While we found that the gene encoding AcrE was conserved across the E. coli types at a level comparable to AcrA and AcrB, AcrF was highly variable. In ST10 alone, five gene variants were identified, but in ST11 there were none. ST11 is a highly infectious E. coli type that includes the Shiga toxin producing O157:H7 that can cause outbreaks of severe infection.
Further work focusing on ST11 identified a highly conserved insertion within the AcrF encoding gene. This insertion when translated into the protein sequence, included 13 amino acids and 2 internal stops codons which truncates the protein roughly half-way through the sequence. We confirmed the presence of this insertion in over 1700 ST11 genome assemblies and confirmed the gene product was unable to restore AcrF function.

It is becoming clearer that efflux systems can vary between isolates and lineages of a single species. I think this is a really important factor to consider as we move forward with the identification of efflux pump inhibitors and novel therapeutics in the fight against antimicrobial resistance. The more we know about the fundamental biology of these pathogens, the better.
For those interested another member of the Blair group, Elizabeth Darby, has recently published work on efflux pump diversity in another human pathogen, Acinetobacter, in Microbial Genomics. You can also read Dr Pugh and Dr Blair's work, 'E. coli ST11 (O157:H7) does not encode a functional AcrF efflux pump' published in Microbiology.