Time to Strep up surveillance?
How many of us have been treated for a sore throat, or a skin infection such as impetigo? Streptococcus pyogenes, also known as group A Streptococcus or StrepA, is often the underlying pathogen. Usually, these extremely common, mild infections are simply uncomfortable and inconvenient. However, other StrepA infections are life-threatening and lead to major disability in survivors; these patients are amongst the most unwell individuals we care for in hospitals. The burden of StrepA disease in lower income settings is even higher, where high rates of StrepA infection is more often associated with life-limiting rheumatic heart disease. These complexities make StrepA a formidable pathogen, and a formidable challenge for researchers and clinicians.
I’m Jenny Hall, an infectious diseases trainee clinician and academic trainee working in the Turner Laboratory in the Molecular Microbiology research cluster in the School of Biosciences and part of the Florey Institute of Infection at University of Sheffield, UK. Our team works to understand the mechanisms behind upsurges in StrepA disease. We use a range of methods to better understand this fascinating pathogen, including whole genome sequencing, analysis of linked epidemiological and clinical data, and phenotypic infection studies using unique 3D tissue-engineered models.
In 2022-23, we saw a dramatic and sudden upsurge of scarlet fever. The impacts of this upsurge were felt widely across society, with the substantial burden on healthcare providers and widespread antibiotic shortages leading to dramatic media attention and issue of emergency clinical guidance. There were also a number of deaths from invasive StrepA disease. There were many hypotheses about potential drivers of this upsurge and there is little doubt that the ‘immunity debt’ associated with the COVID-19 pandemic and the high levels of circulating respiratory viruses, including RSV, played a role. However, the narrative felt incomplete. Were there also bacterial genomic drivers behind this upsurge?
That is the question we sought to address in our recent paper, now published in Microbial Genomics. We did this by collecting and genome sequencing non-invasive swab samples of throat and skin identified as StrepA during routine clinical diagnostic testing in Sheffield, UK, during the upsurge. We compared data with a similar collection from 2016-17 to carry out detailed genomic characterisation.
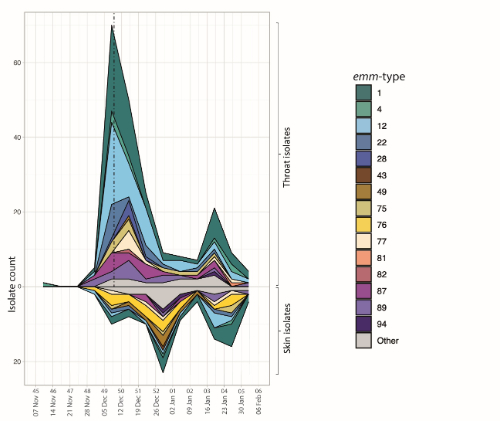
There were a number of interesting findings. Firstly, as well as a rise in throat StrepA isolates, reflecting the national reported rise in scarlet fever, we also identified a rise in skin StrepA during this time. We also recognised that the upsurge did not appear to be driven by a single strain genotype. Whilst emm1 and emm12 were most common genotypes, we also saw a rise in other genotypes such as emm22 and emm49, as well as a fall in other types such as emm89. Furthermore, the distribution of strain types found in skin was different to throat, and interestingly we identified that acapsular strains appeared to be over-represented in throat isolates compared to skin isolates. In contrast, almost all throat isolates were predicted to express high level of toxins, compared to a much lower proportion of skin isolates. These findings highlight elements of tissue tropism in this organism, as well as highlighting for the first time a key role for toxin expression in the pathogenesis of streptococcal throat infections compared to skin infections. Finally, we also identified emergent sublineages in our study, with important findings for potential tissue tropism and future strain diversity.
This work demonstrates the pervasive power of genomics as a tool in understanding bacterial pathogens. Characterising invasive disease alone offers an incomplete picture of StrepA’s overall potential for disease. We argue there is significant potential value in surveillance of non-invasive isolates between upsurges; monitoring this vast reservoir of infection allows us to better understand StrepA tissue tropism, characterise new and emergent sublineages and identify changes in pathogenic potential. We believe that understanding these factors empowers us to understand how non-invasive disease impacts invasive disease. It also allows insight into StrepA overspill into upsurges, allowing us to predict and mitigate the effects.
StrepA continues to pose an incredible challenge at the intersection of clinical medicine, molecular microbiology, public health, and global health. This cross-disciplinary research was a collaboration between researchers from different faculties of the University of Sheffield as well as NHS clinicians and NHS laboratory biomedical scientists. The cross-disciplinary approach to a cross-disciplinary challenge was extremely rewarding. We’re excited to continue our future work in this field, with plans to apply our findings in genotype-to-phenotype infection models, expand our study of tissue tropism, and to explore optimal StrepA surveillance strategies. StrepA is both common and at times devastating; research to tackle StrepA is a critical tool in improving our future health.
Image: iStock/Dr_Microbe
